Due to the characteristics of its dynamic explanatory language, python runs code much slower than java and c + +. Especially when doing scientific computing, the disadvantages of Python are more prominent due to the billions and billions of operations.
The solution is always more difficult than the difficulty. numba is a powerful tool to solve the slow speed of python, which can improve the running speed of Python hundreds of times!
What is numba?
Numba is a JIT compiler that can compile python functions into machine code. The running speed of python code compiled by numba (array operation only) can be close to that of C or FORTRAN language.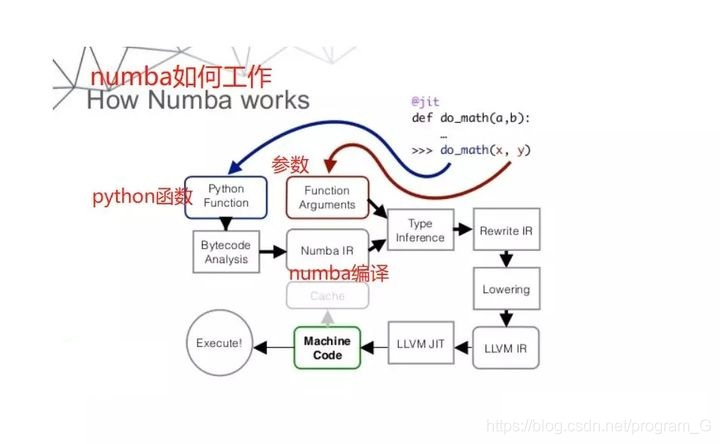
python is slow because it is compiled by CPython. The function of numba is to change a compiler for python.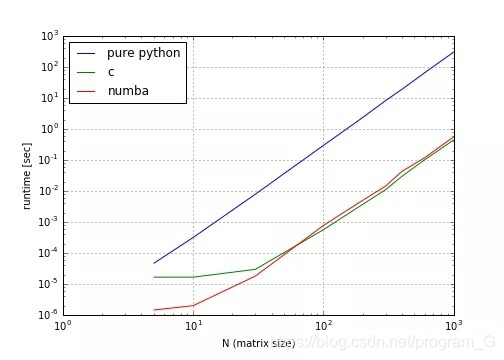
Using numba is very simple. You only need to apply the numba decorator to python functions without changing the original python code. Numba will automatically complete the rest of the work.
import numpy as np
import numba
from numba import jit
@jit(nopython=True) # jit, one of numba decorators
def go_fast(a): # On the first call, the function is compiled into machine code
trace = 0
# Suppose the input variable is a numpy array
for i in range(a.shape[0]): # Numba is good at handling loops
trace += np.tanh(a[i, i])
return a + trace
The above code is a python function to calculate the hyperbolic tangent of each value of numpy array. We use the numba decorator, which compiles this Python function into equivalent machine code, which can greatly reduce the running time.
numba is suitable for scientific computing
Numpy is designed for numpy array oriented computing tasks.
In array oriented computing tasks, data parallelism is natural for accelerators such as GPU. Numba understands numpy array types and uses them to generate efficient compiled code for execution on GPU or multi-core CPU. Special decorators can also create functions that broadcast on numpy arrays like numpy functions.
When to use numba?
- When doing a lot of scientific calculations with numpy arrays
- When using the for loop
Learn to use numba
Step 1: import numpy, numba and their compilers
import numpy as np import numba from numba import jit
Step 2: pass in the numba decorator jit and write the function
# Incoming jit, one of numba decorators
@jit(nopython=True)
def go_fast(a): # On the first call, the function is compiled into machine code
trace = 0
# Suppose the input variable is a numpy array
for i in range(a.shape[0]): # Numba is good at handling loops
trace += np.tanh(a[i, i]) # numba likes numpy functions
return a + trace # numba likes numpy radio
The nopython = True option requires the function to be fully compiled (to completely remove Python interpreter calls), otherwise exceptions will be thrown. These exceptions usually indicate where in the function needs to be modified to achieve better performance than python. It is strongly recommended that you always use nopython = True.
Step 3: pass arguments to the function
# Because the function requires that the parameter passed in is a nunpy array x = np.arange(100).reshape(10, 10) # Execution function go_fast(x)
Step 4: function execution time accelerated by numba
% timeit go_fast(x)
Output: 3.63 µ s ± 156 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)
Step 5: function execution time without numba acceleration
def go_fast(a): # On the first call, the function is compiled into machine code
trace = 0
# Suppose the input variable is a numpy array
for i in range(a.shape[0]): # Numba is good at handling loops
trace += np.tanh(a[i, i]) # numba likes numpy functions
return a + trace # numba likes numpy radio
x = np.arange(100).reshape(10, 10)
%timeit go_fast(x)
Output: 136 µ s ± 1.09 µ s per loop (mean ± std. dev. of 7 runs, 10000 loops each)
conclusion
Under numba acceleration, the code execution time is 3.63 microseconds / cycle. Without numba acceleration, the code execution time is 136 microseconds / cycle, which is 40 times faster than the former.
numba makes python fly
We have compared the speed of python code before and after numba, but this is not the fastest.
This time, instead of using numpy array, we just use for loop to see how much nunba loves for loop!
# Without numba
def t():
x = 0
for i in np.arange(5000):
x += i
return x
%timeit(t())
Output: 408 µ s ± 9.73 µ s per loop (mean ± std. dev. of 7 runs, 1000 loops each)
# Use of numba
@jit(nopython=True)
def t():
x = 0
for i in np.arange(5000):
x += i
return x
%timeit(t())
Output: 1.57 µ s ± 53.8 ns per loop (mean ± std. dev. of 7 runs, 1000000 loops each)
Before and after using numba, it is 408 microseconds / cycle and 1.57 microseconds / cycle respectively, and the speed has been increased by more than 200 times!
epilogue
numba has greatly improved the running speed of Python code, which has greatly promoted the python data analysis ability in the era of big data. For data scientists, this is really a lucky tool!
Of course, numba won't be very helpful to python code other than numpy and for loops. Don't expect numba to help you speed up fetching data from the database. It really can't do that.
Finally, thank you for reading. Each of your likes, comments and sharing is our greatest encouragement. Refill ~
If you have different opinions, welcome to discuss together in the comment area!