In this section, we introduce the method of adding saliency to the box diagram, similar to this:
"Visualization of single factor two-level T inspection box line diagram"

"Visualization of single factor three level T inspection box line diagram"

"Single factor three horizontal column chart"

"Single factor three level line chart"

"Two factor column chart"

"Two factor line chart"

1. Single factor two level
For example, there are differences in plant height between two varieties, and 10 plants are investigated for each variety, which constitutes such test data.
"Analog data:"
set.seed(123)
y1 = rnorm(10) + 5
y2 = rnorm(10) + 15
dd = data.frame(Group = rep(c("A","B"),each=10),y = c(y1,y2))
dd
str(dd)
dd$Group = as.factor(dd$Group)
Data:
> dd Group y 1 A 4.439524 2 A 4.769823 3 A 6.558708 4 A 5.070508 5 A 5.129288 6 A 6.715065 7 A 5.460916 8 A 3.734939 9 A 4.313147 10 A 4.554338 11 B 16.224082 12 B 15.359814 13 B 15.400771 14 B 15.110683 15 B 14.444159 16 B 16.786913 17 B 15.497850 18 B 13.033383 19 B 15.701356 20 B 14.527209
Here, the ggpubr package is used for drawing:
1.1 draw box line diagram
library(ggplot2) library(ggpubr) ggboxplot(dd,x = "Group",y = "y")

Insert picture description here
1.2 add different colors to the box diagram
ggboxplot(dd,x = "Group",y = "y",color = "Group")

1.3 add scatter diagram to box diagram
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter")

1.4 box plot + scatter plot + significance level
Here, the default statistical method is nonparametric statistics Wilcoxon. If you want to use t.test, see the following operation
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") + stat_compare_means()

1.5 t.test is used as the statistical method
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") + stat_compare_means(method = "t.test")

1.6 significance of direct output
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") + stat_compare_means(method = "t.test",label = "p.signif")

2. Single factor and three levels
Two levels can be tested by T. how to test the data of three or more levels?
"Analog data:"
# Construct three level ANOVA
set.seed(123)
y1 = rnorm(10) + 5
y2 = rnorm(10) + 15
y3 = rnorm(10) + 15
dd = data.frame(Group = rep(c("A","B","C"),each=10),y = c(y1,y2,y3))
dd
str(dd)
dd$Group = as.factor(dd$Group)
"The data are as follows:"
> dd Group y 1 A 4.439524 2 A 4.769823 3 A 6.558708 4 A 5.070508 5 A 5.129288 6 A 6.715065 7 A 5.460916 8 A 3.734939 9 A 4.313147 10 A 4.554338 11 B 16.224082 12 B 15.359814 13 B 15.400771 14 B 15.110683 15 B 14.444159 16 B 16.786913 17 B 15.497850 18 B 13.033383 19 B 15.701356 20 B 14.527209 21 C 13.932176 22 C 14.782025 23 C 13.973996 24 C 14.271109 25 C 14.374961 26 C 13.313307 27 C 15.837787 28 C 15.153373 29 C 13.861863 30 C 16.253815
2.1 box diagram + scatter diagram
p = ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") p

2.2 box plot + scatter plot + significance
p + stat_compare_means(method = "anova")
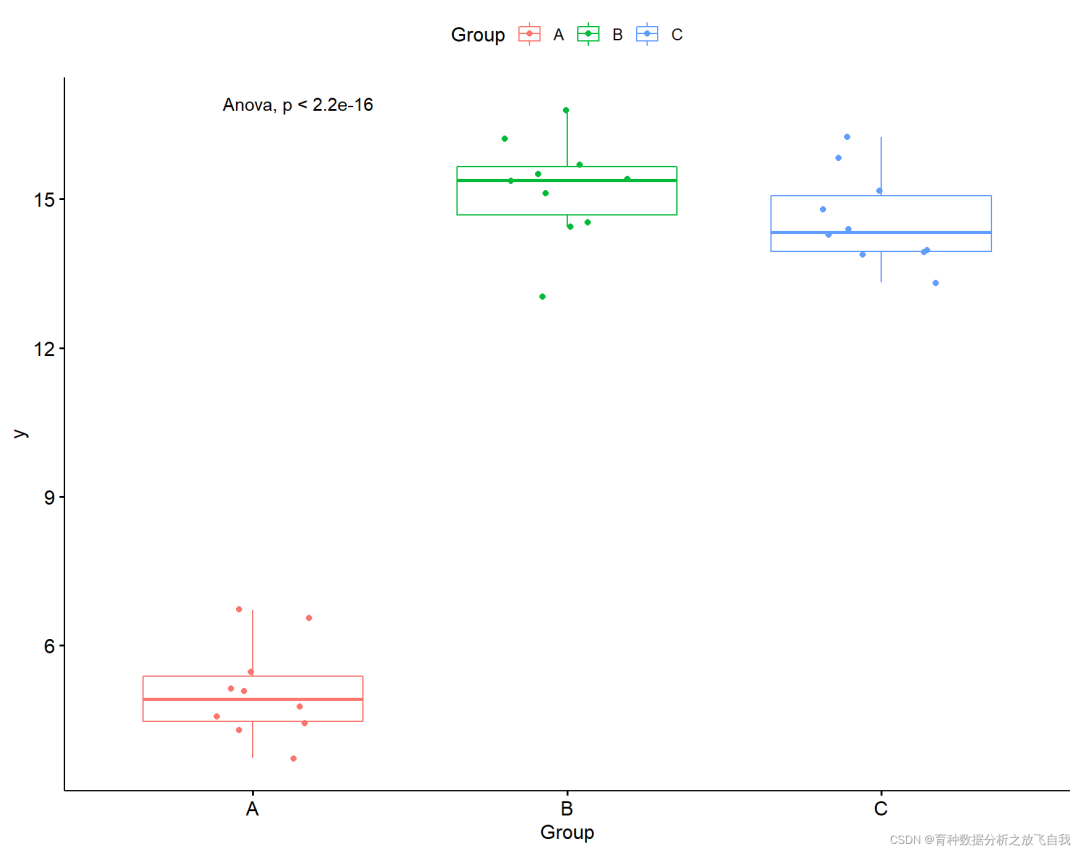
2.3 significance mapping between two
my_comparisons = list( c("A", "B"), c("A", "C"), c("B", "C") )
p + stat_compare_means(comparisons = my_comparisons,
# label = "p.signif",
method = "t.test")
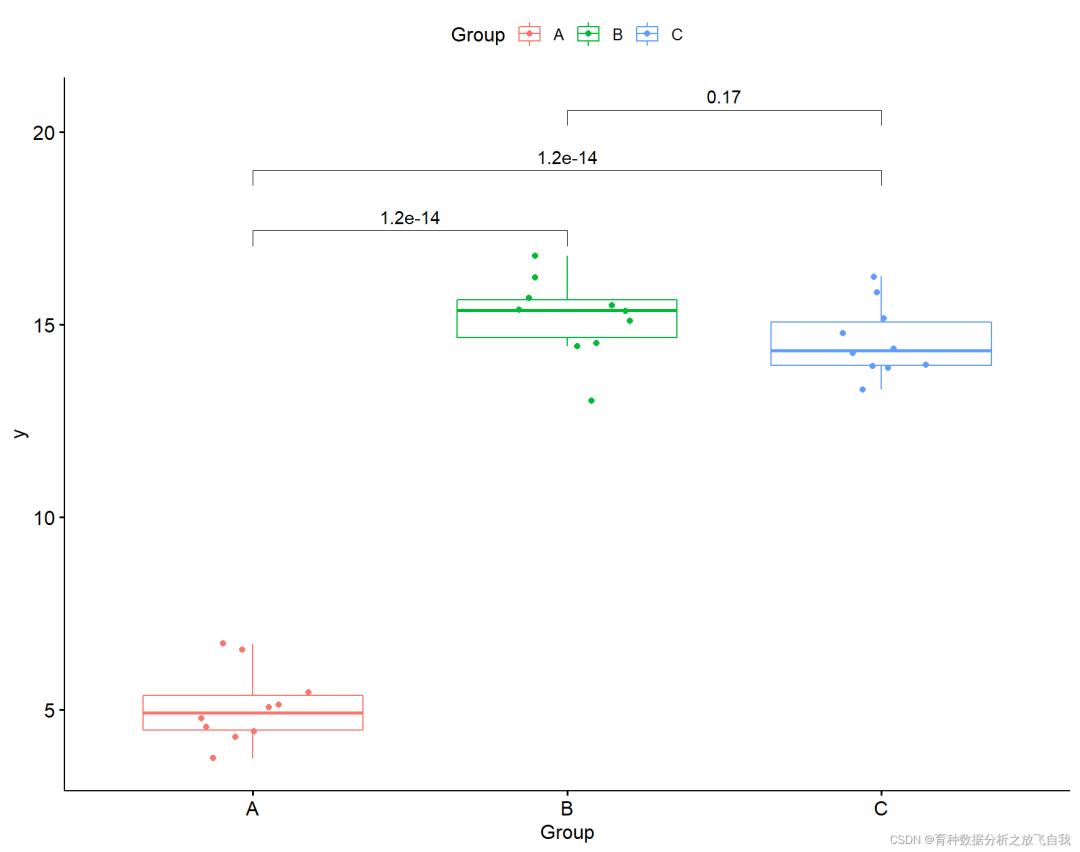
2.4 display significance
p + stat_compare_means(comparisons = my_comparisons,
label = "p.signif",
method = "t.test")

3. Two factor data
"Analog data:"
# Two factor data
set.seed(123)
y1 = rnorm(10) + 5
y2 = rnorm(10) + 8
y3 = rnorm(10) + 7
y4 = rnorm(10) + 15
y5 = rnorm(10) + 18
y6 = rnorm(10) + 17
dd = data.frame(Group1 = rep(c("A","B","C"),each=10),
Group2 = rep(c("X","Y"),each=30),
y = c(y1,y2,y3,y4,y5,y6))
dd
str(dd)
dd$Group1 = as.factor(dd$Group1)
dd$Group2 = as.factor(dd$Group2)
str(dd)
Data preview:
> dd Group1 Group2 y 1 A X 4.439524 2 A X 4.769823 3 A X 6.558708 4 A X 5.070508 5 A X 5.129288 6 A X 6.715065 7 A X 5.460916 8 A X 3.734939 9 A X 4.313147 10 A X 4.554338 11 B X 9.224082 12 B X 8.359814 13 B X 8.400771 14 B X 8.110683 15 B X 7.444159 16 B X 9.786913 17 B X 8.497850 18 B X 6.033383 19 B X 8.701356 20 B X 7.527209 21 C X 5.932176 22 C X 6.782025 23 C X 5.973996 24 C X 6.271109 25 C X 6.374961 26 C X 5.313307 27 C X 7.837787 28 C X 7.153373 29 C X 5.861863 30 C X 8.253815 31 A Y 15.426464 32 A Y 14.704929 33 A Y 15.895126 34 A Y 15.878133 35 A Y 15.821581 36 A Y 15.688640 37 A Y 15.553918 38 A Y 14.938088 39 A Y 14.694037 40 A Y 14.619529 41 B Y 17.305293 42 B Y 17.792083 43 B Y 16.734604 44 B Y 20.168956 45 B Y 19.207962 46 B Y 16.876891 47 B Y 17.597115 48 B Y 17.533345 49 B Y 18.779965 50 B Y 17.916631 51 C Y 17.253319 52 C Y 16.971453 53 C Y 16.957130 54 C Y 18.368602 55 C Y 16.774229 56 C Y 18.516471 57 C Y 15.451247 58 C Y 17.584614 59 C Y 17.123854 60 C Y 17.215942
3.1 draw group box line diagram
p = ggboxplot(dd,x = "Group1",y="y",color = "Group2",
add = "jitter")
p

3.2 increase P value
p + stat_compare_means(aes(group = Group2),method = "t.test")

3.3 is modified to significant results
p + stat_compare_means(aes(group = Group2),method = "t.test",label = "p.signif")

3.4 plot grouped data separately
p = ggboxplot(dd,x = "Group2",y="y",color = "Group1",
add = "jitter",facet.by = "Group1")
p

3.5 group display statistical inspection
p + stat_compare_means(method = "t.test")

3.6 grouping to show significant results
p + stat_compare_means(method = "t.test",label = "p.signif",label.y = 17)

4. Single factor histogram drawing
Histogram + standard error. Before using ggplot2, it takes a long code. Here is a better scheme.
4.1 histogram + standard error
p = ggbarplot(dd,x = "Group1",y = "y",add = "mean_se",color = "Group1") p
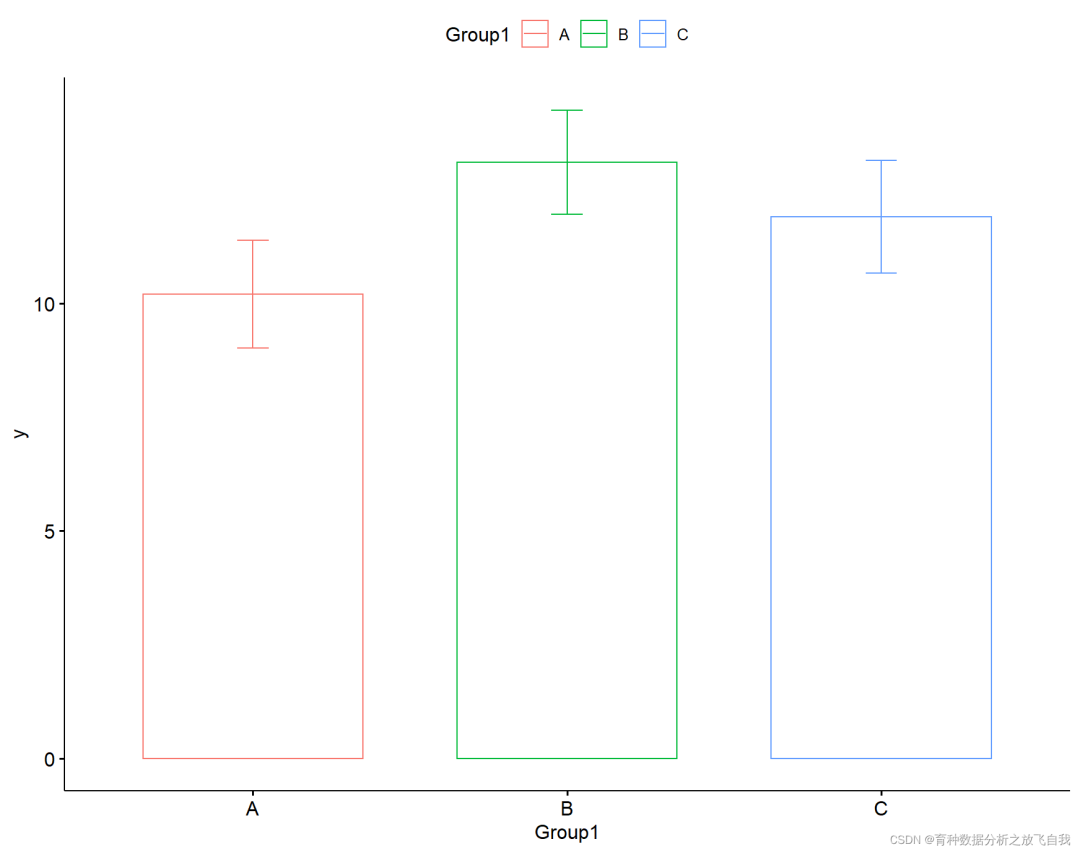
4.2 histogram + standard error + significance
p + stat_compare_means(method = "anova",,label.y = 15)+ stat_compare_means(comparisons = my_comparisons)

5. Single factor line chart drawing
5.1 line chart + standard error
p = ggline(dd,x = "Group1",y = "y",add = "mean_se") p

5.2 line chart + standard error + significance
p + stat_compare_means(method = "anova",,label.y = 15)+ stat_compare_means(comparisons = my_comparisons)

6. Two factor histogram drawing
6.1 histogram + standard error
p = ggbarplot(dd,x = "Group1",y = "y",add = "mean_se",color = "Group2", position = position_dodge(0.8)) p

6.2 histogram + standard error + significance
p + stat_compare_means(aes(group=Group2), label = "p.signif")

7. Drawing of two factor line chart
7.1 line chart + standard error
p = ggline(dd,x = "Group1",y = "y",add = "mean_se",color = "Group2", position = position_dodge(0.8)) p

7.2 line chart + standard error + significance
p + stat_compare_means(aes(group=Group2), label = "p.signif")

8. Code summary
# Welcome to my official account: the release of self analysis of breeding data. Mainly share R language, Python, breeding data analysis, biostatistics, quantitative genetics, mixed linear model, GWAS and GS.
# Build two horizontal t-tests
set.seed(123)
y1 = rnorm(10) + 5
y2 = rnorm(10) + 15
dd = data.frame(Group = rep(c("A","B"),each=10),y = c(y1,y2))
dd
str(dd)
dd$Group = as.factor(dd$Group)
library(ggplot2)
library(ggpubr)
ggboxplot(dd,x = "Group",y = "y")
ggboxplot(dd,x = "Group",y = "y",color = "Group")
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter")
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") +
stat_compare_means()
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") +
stat_compare_means(method = "t.test")
ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter") +
stat_compare_means(method = "t.test",label = "p.signif")
# Construct three level ANOVA
set.seed(123)
y1 = rnorm(10) + 5
y2 = rnorm(10) + 15
y3 = rnorm(10) + 15
dd = data.frame(Group = rep(c("A","B","C"),each=10),y = c(y1,y2,y3))
dd
str(dd)
dd$Group = as.factor(dd$Group)
p = ggboxplot(dd,x = "Group",y = "y",color = "Group",add = "jitter")
p
p + stat_compare_means(method = "anova")
# Perorm pairwise comparisons
# compare_means(y ~ Group, data = dd,method = "anova")
my_comparisons = list( c("A", "B"), c("A", "C"), c("B", "C") )
p + stat_compare_means(comparisons = my_comparisons,
# label = "p.signif",
method = "t.test")
p + stat_compare_means(comparisons = my_comparisons,
label = "p.signif",
method = "t.test")
# Two factor data
set.seed(123)
y1 = rnorm(10) + 5
y2 = rnorm(10) + 8
y3 = rnorm(10) + 7
y4 = rnorm(10) + 15
y5 = rnorm(10) + 18
y6 = rnorm(10) + 17
dd = data.frame(Group1 = rep(c("A","B","C"),each=10),
Group2 = rep(c("X","Y"),each=30),
y = c(y1,y2,y3,y4,y5,y6))
dd
str(dd)
dd$Group1 = as.factor(dd$Group1)
dd$Group2 = as.factor(dd$Group2)
str(dd)
## Group view
p = ggboxplot(dd,x = "Group1",y="y",color = "Group2",
add = "jitter")
p
p + stat_compare_means(aes(group = Group2),method = "t.test")
p + stat_compare_means(aes(group = Group2),method = "t.test",label = "p.signif")
## Group view
p = ggboxplot(dd,x = "Group2",y="y",color = "Group1",
add = "jitter",facet.by = "Group1")
p
p + stat_compare_means(method = "t.test")
p + stat_compare_means(method = "t.test",label = "p.signif",label.y = 17)
# Single grouping
# Three level histogram
p = ggbarplot(dd,x = "Group1",y = "y",add = "mean_se",color = "Group1")
p
p + stat_compare_means(method = "anova",,label.y = 15)+
stat_compare_means(comparisons = my_comparisons)
# Line chart with error
p = ggline(dd,x = "Group1",y = "y",add = "mean_se")
p
p + stat_compare_means(method = "anova",,label.y = 15)+
stat_compare_means(comparisons = my_comparisons)
# Two groups
p = ggbarplot(dd,x = "Group1",y = "y",add = "mean_se",color = "Group2", position = position_dodge(0.8))
p
p + stat_compare_means(aes(group=Group2), label = "p.signif")
# Line chart with error
p = ggline(dd,x = "Group1",y = "y",add = "mean_se",color = "Group2", position = position_dodge(0.8))
p
p + stat_compare_means(aes(group=Group2), label = "p.signif")