Original blog post: minfanphd
Day 61: decision tree (1. Preparation)
Decision tree is the most classical machine learning algorithm Actually, I don't want to add "one" after it It has very good interpretability
- There is only one copy of the data The split data subset only needs to save two arrays: availableInstances and availableAttributes
- There are two construction methods, one is to read in the file to obtain the root node, and the other is to establish the acquisition based on data splitting
- Judge whether the data set is pure, that is, whether all class labels are the same. If so, there is no need to split
- Each node (including non leaf nodes) needs a label, so that it can be classified directly when encountering unprecedented attributes To get this tag, you can vote, that is, getMajorityClass()
- Maximizing information gain is equivalent to minimizing conditional information entropy
- The split data block may be empty. In this case, use an array with length 0 instead of null
Entropy is a measure of uncertainty of random variables. The greater the entropy, the greater the uncertainty of random variables.
Information gain represents the known feature
X
X
The information of X makes the class
Y
Y
The degree to which the uncertainty of Y's information is reduced.
The core of ID3 algorithm is to apply the information gain criterion to select features on each node of the decision tree and construct the decision tree recursively. The specific method is: starting from the root node, calculate the information gain of all possible features for the node, then select the feature with the largest information gain as the feature of the node, establish a child node from the different values of the feature, and then recursively call the above method to the child node to form a decision tree.
package MachineLearning.decisiontree;
import weka.core.*;
import java.io.FileReader;
import java.util.Arrays;
/**
* @description:ID3 Decision tree induction algorithm
* @author: Qing Zhang
* @time: 2021/7/11
*/
public class ID3 {
//data set
Instances dataset;
//Whether the dataset is pure (whether all class labels are the same)
boolean pure;
//Number of classes
int numClasses;
//Available instances. Other instances do not belong to this branch
int[] availableInstances;
//Available properties. Other properties have been selected in the path to the root node
int[] availableAttributes;
//Currently selected split attribute
int splitAttribute;
//Child node
ID3[] children;
//My label. Internal nodes also have labels.
//For example, < outlook = sunny, humidity = high > never appears in the training set,
//However, < humidity = high > is effective in other cases
int label;
//Forecast results, including query and forecast labels
int[] predicts;
//Small blocks cannot be further divided
static int smallBlockThreshold = 3;
/**
* @Description: Constructor
* @Param: [paraFilename]
* @return:
*/
public ID3(String paraFilename) {
dataset = null;
try {
FileReader fileReader = new FileReader(paraFilename);
dataset = new Instances(fileReader);
fileReader.close();
} catch (Exception ee) {
System.out.println("Cannot read the file: " + paraFilename + "\r\n" + ee);
System.exit(0);
}
dataset.setClassIndex(dataset.numAttributes() - 1);
numClasses = dataset.classAttribute().numValues();
availableInstances = new int[dataset.numInstances()];
for (int i = 0; i < availableInstances.length; i++) {
availableInstances[i] = i;
}
availableAttributes = new int[dataset.numAttributes() - 1];
for (int i = 0; i < availableAttributes.length; i++) {
availableAttributes[i] = i;
}
//initialization
children = null;
//Judging labels by voting
label = getMajorityClass(availableInstances);
//Judge whether the instance is pure
pure = pureJudge(availableInstances);
}
/**
* @Description: Constructor
* @Param: [paraDataset, paraAvailableInstances, paraAvailableAttributes]
* @return:
*/
public ID3(Instances paraDataset, int[] paraAvailableInstances, int[] paraAvailableAttributes) {
//Copy its references instead of cloning availableInstances
dataset = paraDataset;
availableInstances = paraAvailableInstances;
availableAttributes = paraAvailableAttributes;
//initialization
children = null;
//Judging labels by voting
label = getMajorityClass(availableInstances);
//Judge whether the instance is pure
pure = pureJudge(availableInstances);
}
/**
* @Description: Judge whether it is pure
* @Param: [paraBlock]
* @return: boolean
*/
public boolean pureJudge(int[] paraBlock) {
pure = true;
for (int i = 1; i < paraBlock.length; i++) {
if (dataset.instance(paraBlock[i]).classValue() != dataset.instance(paraBlock[0])
.classValue()) {
pure = false;
break;
}
}
return pure;
}
/**
* @Description: Calculate the main class of a given block by voting
* @Param: [paraBlock]
* @return: int
*/
public int getMajorityClass(int[] paraBlock) {
int[] tempClassCounts = new int[dataset.numClasses()];
for (int i = 0; i < paraBlock.length; i++) {
tempClassCounts[(int) dataset.instance(paraBlock[i]).classValue()]++;
}
int resultMajorityClass = -1;
int tempMaxCount = -1;
for (int i = 0; i < tempClassCounts.length; i++) {
if (tempMaxCount < tempClassCounts[i]) {
resultMajorityClass = i;
tempMaxCount = tempClassCounts[i];
}
}
return resultMajorityClass;
}
/**
* @Description: Select optimal attribute
* @Param: []
* @return: int
*/
public int selectBestAttribute() {
splitAttribute = -1;
double tempMinimalEntropy = 10000;
double tempEntropy;
//The minimum conditional entropy is selected as the optimal attribute
for (int i = 0; i < availableAttributes.length; i++) {
tempEntropy = conditionalEntropy(availableAttributes[i]);
if (tempMinimalEntropy > tempEntropy) {
tempMinimalEntropy = tempEntropy;
splitAttribute = availableAttributes[i];
}
}
return splitAttribute;
}
/**
* @Description: Calculating conditional entropy
* @Param: [paraAttribute]
* @return: double
*/
public double conditionalEntropy(int paraAttribute) {
// Step 1. Statistics, statistics of the category distribution under this feature
//Number of categories in the dataset
int tempNumClasses = dataset.numClasses();
//Number of categories in which the current attribute is classified
int tempNumValues = dataset.attribute(paraAttribute).numValues();
int tempNumInstances = availableInstances.length;
double[] tempValueCounts = new double[tempNumValues];
double[][] tempCountMatrix = new double[tempNumValues][tempNumClasses];
int tempClass, tempValue;
for (int i = 0; i < tempNumInstances; i++) {
tempClass = (int) dataset.instance(availableInstances[i]).classValue();
tempValue = (int) dataset.instance(availableInstances[i]).value(paraAttribute);
tempValueCounts[tempValue]++;
tempCountMatrix[tempValue][tempClass]++;
}
// Step 2.
double resultEntropy = 0;
double tempEntropy, tempFraction;
for (int i = 0; i < tempNumValues; i++) {
if (tempValueCounts[i] == 0) {
continue;
}
tempEntropy = 0;
for (int j = 0; j < tempNumClasses; j++) {
//The probability of one of the cases under this characteristic
tempFraction = tempCountMatrix[i][j] / tempValueCounts[i];
if (tempFraction == 0) {
continue;
}
tempEntropy += -tempFraction * Math.log(tempFraction);
}
resultEntropy += tempValueCounts[i] / tempNumInstances * tempEntropy;
}
return resultEntropy;
}
/**
* @Description: Split data according to given attributes
* @Param: [paraAttribute]
* @return: int[][]
*/
public int[][] splitData(int paraAttribute) {
int tempNumValues = dataset.attribute(paraAttribute).numValues();
// System.out.println("Dataset " + dataset + "\r\n");
// System.out.println("Attribute " + paraAttribute + " has " +
// tempNumValues + " values.\r\n");
int[][] resultBlocks = new int[tempNumValues][];
int[] tempSizes = new int[tempNumValues];
//The first scan is used to calculate the size of each block
int tempValue;
for (int i = 0; i < availableInstances.length; i++) {
tempValue = (int) dataset.instance(availableInstances[i]).value(paraAttribute);
tempSizes[tempValue]++;
}
//Allocate space
for (int i = 0; i < tempNumValues; i++) {
resultBlocks[i] = new int[tempSizes[i]];
}
//In the second round of scanning, fill tempSizes to zero, and then divide the instances into corresponding blocks in turn
Arrays.fill(tempSizes, 0);
for (int i = 0; i < availableInstances.length; i++) {
tempValue = (int) dataset.instance(availableInstances[i]).value(paraAttribute);
//Copy data
resultBlocks[tempValue][tempSizes[tempValue]] = availableInstances[i];
tempSizes[tempValue]++;
}
return resultBlocks;
}
/**
* @Description: Establish regression tree
* @Param: []
* @return: void
*/
public void buildTree() {
//If the available instances are pure, there is no need to divide the tree
if (pureJudge(availableInstances)) {
return;
}
//If the remaining instances are less than the block threshold, the partition can also be stopped
if (availableInstances.length <= smallBlockThreshold) {
return;
}
selectBestAttribute();
int[][] tempSubBlocks = splitData(splitAttribute);
children = new ID3[tempSubBlocks.length];
//Construct the remaining attribute set
int[] tempRemainingAttributes = new int[availableAttributes.length - 1];
for (int i = 0; i < availableAttributes.length; i++) {
if (availableAttributes[i] < splitAttribute) {
tempRemainingAttributes[i] = availableAttributes[i];
} else if (availableAttributes[i] > splitAttribute) {
tempRemainingAttributes[i - 1] = availableAttributes[i];
}
}
// Build son
for (int i = 0; i < children.length; i++) {
if ((tempSubBlocks[i] == null) || (tempSubBlocks[i].length == 0)) {
children[i] = null;
continue;
} else {
// System.out.println("Building children #" + i + " with
// instances " + Arrays.toString(tempSubBlocks[i]));
children[i] = new ID3(dataset, tempSubBlocks[i], tempRemainingAttributes);
//Important code: perform this operation recursively
children[i].buildTree();
}
}
}
/**
* @Description: Judge instance category
* @Param: [paraInstance]
* @return: int
*/
public int classify(Instance paraInstance) {
if (children == null) {
return label;
}
ID3 tempChild = children[(int) paraInstance.value(splitAttribute)];
if (tempChild == null) {
return label;
}
return tempChild.classify(paraInstance);
}
/**
* @Description: Test on test set
* @Param: [paraDataset]
* @return: double
*/
public double test(Instances paraDataset) {
double tempCorrect = 0;
for (int i = 0; i < paraDataset.numInstances(); i++) {
if (classify(paraDataset.instance(i)) == (int) paraDataset.instance(i).classValue()) {
tempCorrect++;
}
}
return tempCorrect / paraDataset.numInstances();
}
/**
* @Description: Test on training set
* @Param: []
* @return: double
*/
public double selfTest() {
return test(dataset);
}
public String toString() {
String resultString = "";
String tempAttributeName = dataset.attribute(splitAttribute).name();
if (children == null) {
resultString += "class = " + label;
} else {
for (int i = 0; i < children.length; i++) {
if (children[i] == null) {
resultString += tempAttributeName + " = "
+ dataset.attribute(splitAttribute).value(i) + ":" + "class = " + label
+ "\r\n";
} else {
resultString += tempAttributeName + " = "
+ dataset.attribute(splitAttribute).value(i) + ":" + children[i]
+ "\r\n";
}
}
}
return resultString;
}
/**
* @Description: Test this class
* @Param: []
* @return: void
*/
public static void id3Test() {
ID3 tempID3 = new ID3("F:\\graduate student\\Yan 0\\study\\Java_Study\\data_set\\weather.arff");
// ID3 tempID3 = new ID3("D:/data/mushroom.arff");
ID3.smallBlockThreshold = 3;
tempID3.buildTree();
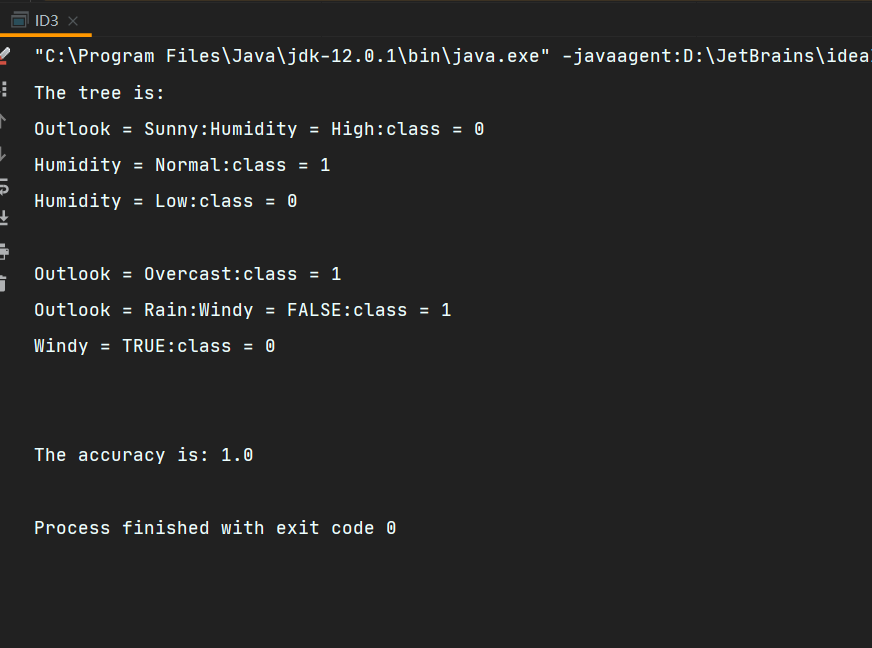
System.out.println("The tree is: \r\n" + tempID3);
double tempAccuracy = tempID3.selfTest();
System.out.println("The accuracy is: " + tempAccuracy);
}
public static void main(String[] args) {
id3Test();
}
}