Refer to the official website: Scipy.
Fixed point problem
The problem closely related to finding the zero point of a function is to find the fixed point of a function. The fixed point of a function refers to the point returned when the function is evaluated: g ( x ) = x g(x)=x g(x)=x. Obviously, this is f ( x ) = g ( x ) − x f(x)=g(x)-x The zero point problem of f(x)=g(x) − X. fixed_point provides a simple iterative method, which uses Aitkens sequence acceleration to estimate g ( x ) g(x) The fixed point of g(x).
from scipy import optimize import numpy as np def func(x, c1, c2): return np.sqrt(c1/(x+c2)) c1 = np.array([10,12.]) c2 = np.array([3, 5.]) ans = optimize.fixed_point(func, [1.2, 1.3], args=(c1,c2)) print(ans)
The result is:
[1.4920333 1.37228132]
Root finding of univariate equation
For example:
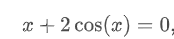
import numpy as np
from scipy.optimize import root
def func(x):
return x + 2 * np.cos(x)
sol = root(func, 0.3)
print(sol.x)
The results are:
[-1.02986653]
Root finding of multivariable equation (method = 'lm')
For example:
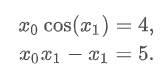
import numpy as np
from scipy.optimize import root
def func2(x):
f = [x[0] * np.cos(x[1]) - 4,
x[1]*x[0] - x[1] - 5]
df = np.array([[np.cos(x[1]), -x[0] * np.sin(x[1])],
[x[1], x[0] - 1]])
return f, df
sol = root(func2, [1, 1], jac=True, method='lm')
print(sol.x)
The results are:
[6.50409711 0.90841421]
Large scale root problem
hybr and lm cannot solve large-scale problems because they have to calculate n × N Jacobian matrix, with the increase of variables, the amount of calculation will be greater and greater.
For example, consider the following problems: we need to solve the following differential equations:
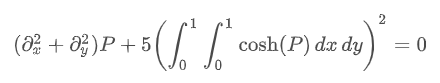
The limiting conditions are: the upper edge meets
P
(
x
,
1
)
=
1
P(x,1)=1
P(x,1)=1, satisfied on the boundary
P
=
0
P=0
P=0. This can be achieved by approximating the continuous function p with the values on the grid (personal understanding is the finite difference method for fluid)
P
n
,
m
≈
P
(
n
h
,
m
h
)
P_{n,m}≈P(nh,mh)
Pn,m≈P(nh,mh). Then the derivative and integral can be approximated; For example:
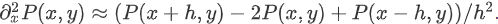
In this way, large solvers can be used to solve: such as krylov, broyden2 and anderson. These methods use the so-called inexact Newton method, which does not accurately calculate the Jacobian matrix, but forms an approximate value.
import numpy as np
from scipy.optimize import root
from numpy import cosh, zeros_like, mgrid, zeros
# parameters
nx, ny = 75, 75
hx, hy = 1./(nx-1), 1./(ny-1)
P_left, P_right = 0, 0
P_top, P_bottom = 1, 0
def residual(P):
d2x = zeros_like(P)
d2y = zeros_like(P)
d2x[1:-1] = (P[2:] - 2*P[1:-1] + P[:-2]) / hx/hx
d2x[0] = (P[1] - 2*P[0] + P_left)/hx/hx
d2x[-1] = (P_right - 2*P[-1] + P[-2])/hx/hx
d2y[:,1:-1] = (P[:,2:] - 2*P[:,1:-1] + P[:,:-2])/hy/hy
d2y[:,0] = (P[:,1] - 2*P[:,0] + P_bottom)/hy/hy
d2y[:,-1] = (P_top - 2*P[:,-1] + P[:,-2])/hy/hy
return d2x + d2y + 5*cosh(P).mean()**2
# solve
guess = zeros((nx, ny), float)
sol = root(residual, guess, method='krylov', options={'disp': True})
#sol = root(residual, guess, method='broyden2', options={'disp': True, 'max_rank': 50})
#sol = root(residual, guess, method='anderson', options={'disp': True, 'M': 10})
print('Residual: %g' % abs(residual(sol.x)).max())
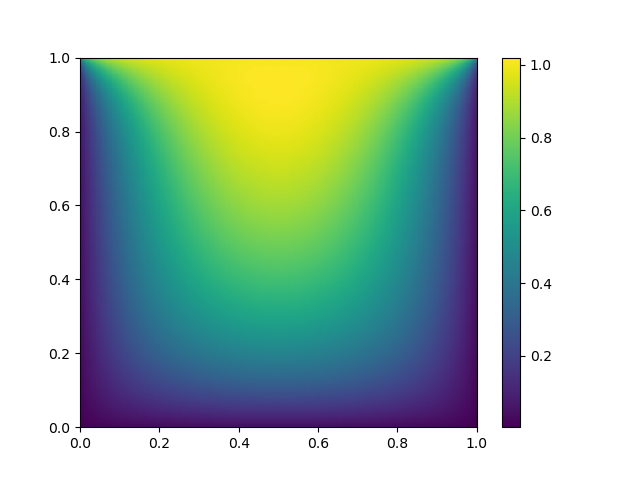
# visualize
import matplotlib.pyplot as plt
x, y = mgrid[0:1:(nx*1j), 0:1:(ny*1j)]
plt.pcolormesh(x, y, sol.x, shading='gouraud')
plt.colorbar()
plt.show()
The result is:
0: |F(x)| = 40.1231; step 1 1: |F(x)| = 16.821; step 1 2: |F(x)| = 5.69025; step 1 3: |F(x)| = 1.41135; step 1 4: |F(x)| = 0.0386739; step 1 5: |F(x)| = 0.00171956; step 1 6: |F(x)| = 0.000132096; step 1 7: |F(x)| = 6.2608e-06; step 1 8: |F(x)| = 3.69188e-07; step 1 Residual: 3.69188e-07
Acceleration method
For krylov method, most of its time is used to calculate the inverse of Jacobian matrix. If you have an approximation of the inverse matrix
M
≈
J
−
1
M≈J^{-1}
M ≈ J − 1, then you can use it to preprocess the linear inversion problem.
We're not going to solve it
J
s
=
y
Js=y
Js=y, but solve it
M
J
s
=
M
y
MJs=My
MJs=My. Because the matrix is "closer" to the identity matrix than before, the Krylov method should be easier to deal with this equation.
Matrix M can be passed to Krylov method as an option options['jac_options']['inner_M '].
For the problem in the previous section, we notice that the function to be solved consists of two parts: the first part is Laplace operator and the second is integral. In fact, we can easily calculate the Jacobian coefficient corresponding to the Laplacian part:

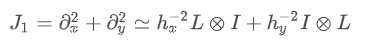
J
1
J_1
J1. You can use SciPy sparse. linalg. Splu (or approximate with scipy.sparse.linalg.splu). So we use
M
≈
J
1
−
1
M≈J^{-1}_1
M ≈ J1 − 1 ≈ is pretreated to accelerate the calculation.
import numpy as np
from scipy.optimize import root
from scipy.sparse import spdiags, kron
from scipy.sparse.linalg import spilu, LinearOperator
from numpy import cosh, zeros_like, mgrid, zeros, eye
# parameters
nx, ny = 75, 75
hx, hy = 1./(nx-1), 1./(ny-1)
P_left, P_right = 0, 0
P_top, P_bottom = 1, 0
def get_preconditioner():
"""Compute the preconditioner M"""
diags_x = zeros((3, nx))
diags_x[0,:] = 1/hx/hx
diags_x[1,:] = -2/hx/hx
diags_x[2,:] = 1/hx/hx
Lx = spdiags(diags_x, [-1,0,1], nx, nx)
diags_y = zeros((3, ny))
diags_y[0,:] = 1/hy/hy
diags_y[1,:] = -2/hy/hy
diags_y[2,:] = 1/hy/hy
Ly = spdiags(diags_y, [-1,0,1], ny, ny)
J1 = kron(Lx, eye(ny)) + kron(eye(nx), Ly)
# Now we have the matrix `J_1`. We need to find its inverse `M` --
# however, since an approximate inverse is enough, we can use
# the *incomplete LU* decomposition
J1_ilu = spilu(J1)
# This returns an object with a method .solve() that evaluates
# the corresponding matrix-vector product. We need to wrap it into
# a LinearOperator before it can be passed to the Krylov methods:
M = LinearOperator(shape=(nx*ny, nx*ny), matvec=J1_ilu.solve)
return M
def solve(preconditioning=True):
"""Compute the solution"""
count = [0]
def residual(P):
count[0] += 1
d2x = zeros_like(P)
d2y = zeros_like(P)
d2x[1:-1] = (P[2:] - 2*P[1:-1] + P[:-2])/hx/hx
d2x[0] = (P[1] - 2*P[0] + P_left)/hx/hx
d2x[-1] = (P_right - 2*P[-1] + P[-2])/hx/hx
d2y[:,1:-1] = (P[:,2:] - 2*P[:,1:-1] + P[:,:-2])/hy/hy
d2y[:,0] = (P[:,1] - 2*P[:,0] + P_bottom)/hy/hy
d2y[:,-1] = (P_top - 2*P[:,-1] + P[:,-2])/hy/hy
return d2x + d2y + 5*cosh(P).mean()**2
# preconditioner
if preconditioning:
M = get_preconditioner()
else:
M = None
# solve
guess = zeros((nx, ny), float)
sol = root(residual, guess, method='krylov',
options={'disp': True,
'jac_options': {'inner_M': M}})
print('Residual', abs(residual(sol.x)).max())
print('Evaluations', count[0])
return sol.x
def main():
sol = solve(preconditioning=True)
# visualize
import matplotlib.pyplot as plt
x, y = mgrid[0:1:(nx*1j), 0:1:(ny*1j)]
plt.clf()
plt.pcolor(x, y, sol)
plt.clim(0, 1)
plt.colorbar()
plt.show()
if __name__ == "__main__":
main()
Good preprocessing may be crucial, and it can even determine whether the problem can be solved in practice.