We have shared the notes of cell Ranger process many times in the single cell world. You can go and learn by yourself, as follows:
- Single cell combat (I) data download
- Single cell combat (II) precautions before using cell ranger
- Single cell combat (III) preliminary study on the use of Cell Ranger
- Single cell combat (IV) overview of Cell Ranger process
- Single cell combat (V) understanding the results of cell Ranger count
Because this process actually requires the fastq file of 10X single cell transcriptome, and the naming is regular!
If your samples are distributed to multiple libraries and flowcell s, there will be 84 fastq files in one sample. I happen to see the data in a document. The study was published in the journal Nature Communications in March 2021, with the title of "time resolved single cell analysis of BRCA1 associated matrix tumorigenesis reviews aberrant differentiation of luminal generators". The link is: https://www.nature.com/articles/s41467-021-21783-3
The sample shown below is SIGAA11. There are 84 fastq files. If you carefully observe the names of these 84 fastq files, you will find the rule. If the underline is the separator, then
- The second column is S37 to S40
- The third column is L003 to L009
- The fifth column is R1, R1 and I1
There are 4x7x3=84 fastq files in total.
Of course, not every 10X sample has 84 fastq files. Even in most cases, only three files or two files can run the cell Ranger process we shared earlier.
The next special case I want to introduce is that there are 44 fastq files, but there is no way to correspond to 10x samples:
SRR15860129 week 11 MMTV-PyMT Week11-1 SRR15860128 week 11 MMTV-PyMT Week11-2 SRR15860127 week 11 MMTV-PyMT Week11-3 SRR15860126 week 11 MMTV-PyMT Week11-4 SRR15860125 week 17 MMTV-PyMT Week17-1 SRR15860124 week 17 MMTV-PyMT Week17-2 SRR15860123 week 17 MMTV-PyMT Week17-3 SRR15860122 week 17 MMTV-PyMT Week17-4 SRR15860120 week 17 MMTV-PyMT Week17-5 SRR15860119 week 17 MMTV-PyMT Week17-6 SRR15860118 week 17 MMTV-PyMT Week17-7 SRR15860117 week 17 MMTV-PyMT Week17-8 SRR15860155 week 7 MMTV-PyMT Week7-1 SRR15860154 week 7 MMTV-PyMT Week7-2 SRR15860143 week 7 MMTV-PyMT Week7-3 SRR15860132 week 7 MMTV-PyMT Week7-4 SRR15860121 week 9 MMTV-PyMT Week9-1 SRR15860150 week 9 MMTV-PyMT Week9-10 SRR15860149 week 9 MMTV-PyMT Week9-11 SRR15860148 week 9 MMTV-PyMT Week9-12 SRR15860147 week 9 MMTV-PyMT Week9-13 SRR15860146 week 9 MMTV-PyMT Week9-14 SRR15860145 week 9 MMTV-PyMT Week9-15 SRR15860144 week 9 MMTV-PyMT Week9-16 SRR15860142 week 9 MMTV-PyMT Week9-17 SRR15860141 week 9 MMTV-PyMT Week9-18 SRR15860140 week 9 MMTV-PyMT Week9-19 SRR15860116 week 9 MMTV-PyMT Week9-2 SRR15860139 week 9 MMTV-PyMT Week9-20 SRR15860138 week 9 MMTV-PyMT Week9-21 SRR15860137 week 9 MMTV-PyMT Week9-22 SRR15860136 week 9 MMTV-PyMT Week9-23 SRR15860135 week 9 MMTV-PyMT Week9-24 SRR15860134 week 9 MMTV-PyMT Week9-25 SRR15860133 week 9 MMTV-PyMT Week9-26 SRR15860131 week 9 MMTV-PyMT Week9-27 SRR15860130 week 9 MMTV-PyMT Week9-28 SRR15860115 week 9 MMTV-PyMT Week9-3 SRR15860114 week 9 MMTV-PyMT Week9-4 SRR15860113 week 9 MMTV-PyMT Week9-5 SRR15860112 week 9 MMTV-PyMT Week9-6 SRR15860153 week 9 MMTV-PyMT Week9-7 SRR15860152 week 9 MMTV-PyMT Week9-8 SRR15860151 week 9 MMTV-PyMT Week9-9
You can see that according to the second column, the 44 fastq files should belong to four 10x samples, so you can run the cellranger process four times. However, if you change your name and run the cellranger process, you will report an error.
In order to solve this problem, I first run the cell Ranger process independently for the 44 fastq files, and the results are read in batches. The code is as follows:
rm(list=ls())
library(data.table)
dir='/home/data/jmzeng/scRNA/mice-4-stage/tmp/matrix'
# Below this folder are the matrix folders of 44 fastq files running cell Ranger processes independently
samples=list.files( dir )
samples
sceList = lapply(samples,function(pro){
# pro=samples[1]
folder=file.path( dir ,pro)
print(pro)
print(folder)
print(list.files(folder))
sce=CreateSeuratObject(counts = Read10X(folder),
project = pro ,
min.cells = 5,
min.features = 300)
return(sce)
})
names(sceList)
If these 44 fastq files belong to different 10x samples, their cell barcodes should theoretically intersect when they go through the cell Ranger process independently, so I made a statistics.
out = do.call(rbind,
lapply(1:length(sceList), function(i){
do.call(rbind,
lapply(1:length(sceList), function(j){
sample1 <- as.data.frame(sceList[[i]]@meta.data)
sample1_barcodes <- as.numeric(length(sample1$orig.ident))
sample2 <- as.data.frame(sceList[[j]]@meta.data)
sample2_barcodes <- as.numeric(length(sample2$orig.ident))
both <- intersect(rownames(sample1),rownames(sample2))
both_barcodes <- as.numeric(length(both))
rate <- as.numeric(both_barcodes/(sample1_barcodes+sample2_barcodes))
c(sample1_name=as.character(unique(sample1$orig.ident)),
sample1_barcodes=sample1_barcodes,
sample2_name=as.character(unique(sample2$orig.ident)),
sample2_barcodes=sample2_barcodes,
rate=rate)
}))
}))
save(out,file = 'out.Rdata')
The single cell matrices of these 44 fastq files are combined and compared:
> head(out)
sample1_name sample1_barcodes sample2_name sample2_barcodes rate
[1,] "SRR15860112" "2197" "SRR15860112" "2197" "0.5"
[2,] "SRR15860112" "2197" "SRR15860113" "1923" "0.465776699029126"
[3,] "SRR15860112" "2197" "SRR15860114" "2746" "0.00101153145862836"
[4,] "SRR15860112" "2197" "SRR15860115" "2768" "0.00100704934541793"
[5,] "SRR15860112" "2197" "SRR15860116" "2811" "0.000998402555910543"
[6,] "SRR15860112" "2197" "SRR15860117" "5100" "0.00315198026586268"
If the coincidence degree of barcodes of the paired cells of the two results is very high, it indicates that it is the same sample. As shown below, SRR15860112 and SRR15860113 are the same sample.
library(tidyr) outs <- as.data.frame(out[,-c(2,4)] ) library(reshape2) out_wide <- dcast(outs,sample1_name~sample2_name) name=out_wide[,1] out_wide=out_wide[,-1] out_wide <- apply(out_wide,2,as.numeric) rownames(out_wide) <- name library(pheatmap)
As follows:
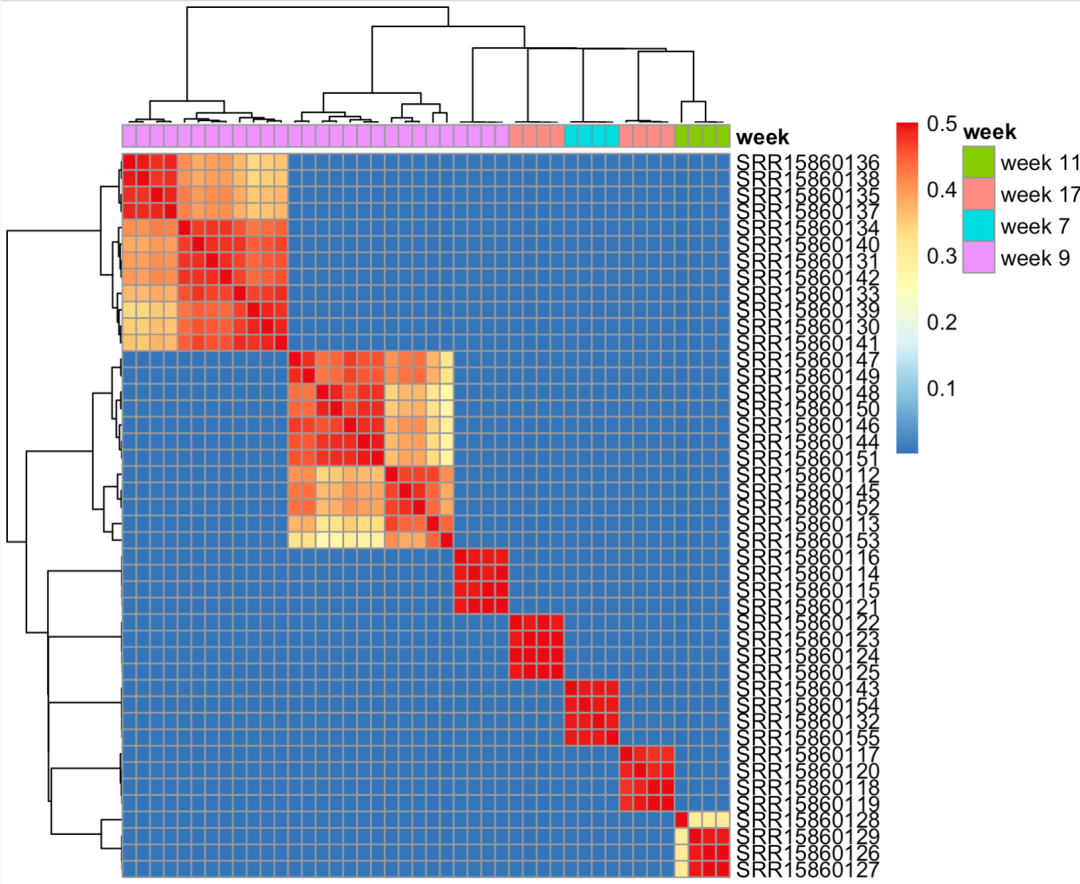
The coincidence degree of cell barcodes in some samples is very high
You can see that our 44 fastq files should belong to 7 samples, so I output the following file list:
head(tmp)
srr hc age
SRR15860126 SRR15860126 5 week 11
SRR15860127 SRR15860127 5 week 11
SRR15860128 SRR15860128 5 week 11
SRR15860129 SRR15860129 5 week 11
SRR15860117 SRR15860117 3 week 17
SRR15860118 SRR15860118 3 week 17
Then change your name:
1_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860146_1.fastq.gz 1_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860146_2.fastq.gz 1_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860147_1.fastq.gz 1_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860147_2.fastq.gz 1_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860148_1.fastq.gz 1_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860148_2.fastq.gz 1_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860149_1.fastq.gz 1_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860149_2.fastq.gz 1_S1_L005_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860150_1.fastq.gz 1_S1_L005_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860150_2.fastq.gz 1_S1_L006_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860151_1.fastq.gz 1_S1_L006_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860151_2.fastq.gz 1_S1_L007_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860152_1.fastq.gz 1_S1_L007_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860152_2.fastq.gz 1_S1_L008_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860153_1.fastq.gz 1_S1_L008_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860153_2.fastq.gz 1_S2_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860112_1.fastq.gz 1_S2_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860112_2.fastq.gz 1_S2_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860113_1.fastq.gz 1_S2_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860113_2.fastq.gz 1_S2_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860144_1.fastq.gz 1_S2_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860144_2.fastq.gz 1_S2_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860145_1.fastq.gz 1_S2_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860145_2.fastq.gz 2_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860114_1.fastq.gz 2_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860114_2.fastq.gz 2_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860115_1.fastq.gz 2_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860115_2.fastq.gz 2_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860116_1.fastq.gz 2_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860116_2.fastq.gz 2_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860121_1.fastq.gz 2_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860121_2.fastq.gz 3_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860117_1.fastq.gz 3_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860117_2.fastq.gz 3_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860118_1.fastq.gz 3_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860118_2.fastq.gz 3_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860119_1.fastq.gz 3_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860119_2.fastq.gz 3_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860120_1.fastq.gz 3_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860120_2.fastq.gz 4_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860122_1.fastq.gz 4_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860122_2.fastq.gz 4_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860123_1.fastq.gz 4_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860123_2.fastq.gz 4_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860124_1.fastq.gz 4_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860124_2.fastq.gz 4_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860125_1.fastq.gz 4_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860125_2.fastq.gz 5_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860126_1.fastq.gz 5_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860126_2.fastq.gz 5_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860127_1.fastq.gz 5_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860127_2.fastq.gz 5_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860128_1.fastq.gz 5_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860128_2.fastq.gz 5_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860129_1.fastq.gz 5_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860129_2.fastq.gz 6_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860130_1.fastq.gz 6_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860130_2.fastq.gz 6_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860131_1.fastq.gz 6_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860131_2.fastq.gz 6_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860133_1.fastq.gz 6_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860133_2.fastq.gz 6_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860134_1.fastq.gz 6_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860134_2.fastq.gz 6_S1_L005_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860135_1.fastq.gz 6_S1_L005_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860135_2.fastq.gz 6_S1_L006_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860136_1.fastq.gz 6_S1_L006_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860136_2.fastq.gz 6_S1_L007_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860137_1.fastq.gz 6_S1_L007_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860137_2.fastq.gz 6_S1_L008_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860138_1.fastq.gz 6_S1_L008_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860138_2.fastq.gz 6_S2_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860139_1.fastq.gz 6_S2_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860139_2.fastq.gz 6_S2_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860140_1.fastq.gz 6_S2_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860140_2.fastq.gz 6_S2_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860141_1.fastq.gz 6_S2_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860141_2.fastq.gz 6_S2_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860142_1.fastq.gz 6_S2_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860142_2.fastq.gz 7_S1_L001_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860132_1.fastq.gz 7_S1_L001_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860132_2.fastq.gz 7_S1_L002_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860143_1.fastq.gz 7_S1_L002_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860143_2.fastq.gz 7_S1_L003_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860154_1.fastq.gz 7_S1_L003_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860154_2.fastq.gz 7_S1_L004_R1_001.fastq.gz -> /home/PRJNA762594/SRR15860155_1.fastq.gz 7_S1_L004_R2_001.fastq.gz -> /home/PRJNA762594/SRR15860155_2.fastq.gz
Then, such a file can run the cell Ranger process.
Finally, the dimension reduction clustering and biological naming are as follows:
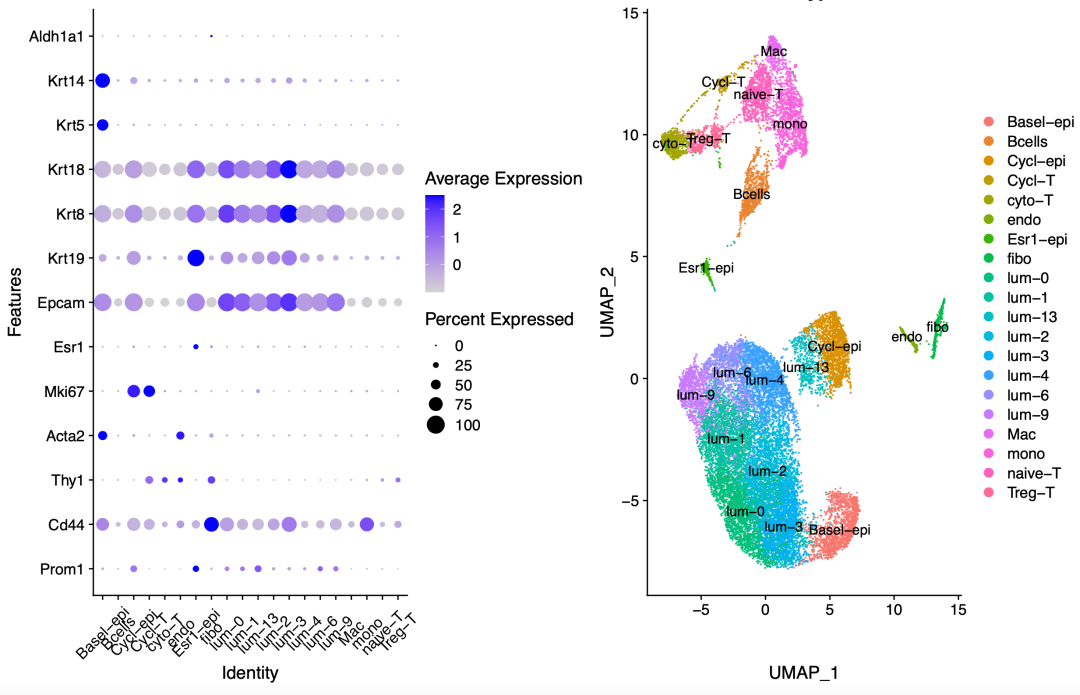
image-20220223150214864
For routine single cell analysis, please refer to the previous example: Single cell clustering and clustering annotation that everyone can learn , we demonstrate the first level of clustering. If you don't have a basic understanding of single-cell data analysis, you can see basic 10:
- 01. Upstream analysis process
- 02. How many samples of the subject and how much sequencing data
- 03. Filter unqualified cells and genes (data quality control is very important)
- 04. Filter mitochondrial ribosomal genes
- 05. Remove cellular and genetic effects
- 06. Dimensionality reduction clustering of single cell transcriptome data
- 07. Cell subpopulation annotation for single cell transcriptome data processing
- 08. Group the obtained subgroups more carefully
- 09. Comparison of cell subsets in single cell transcriptome data processing