
1 Introduction to causalnex
1.1 installation
pip install causalnex
github:https://github.com/quantumblacklabs/causalnex
Document link: https://causalnex.readthedocs.io/en/latest/02_getting_started/02_install.html
CausalNex is a Python library that uses Bayesian networks to combine machine learning and domain expertise for causal reasoning.
You can use CausalNex to reveal structural relationships in data, understand complex distributions, and observe the effects of potential interventions.
The CausalNex library has the following features:
- Adopt the most advanced structural learning method, DAG with no teachers, to understand the conditional dependence between variables
- Allow domain knowledge to extend model relationships
- A prediction model based on structural relationship is established
- Understanding probability model
- Evaluate model quality with standard statistical inspection
- Visualization simplifies the understanding of causality
- The impact of interventions was analyzed using calculus
Compared with the traditional machine learning methods based on pattern recognition and correlation analysis, Bayesian network is more intuitive to describe causality.
If domain expertise can be easily coded or added to the graph model, causality will be more accurate.
Graphical models can then be used to assess the impact of changes on potential characteristics, i.e. counterfactual analysis, and to determine the correct intervention.
According to experience, data scientists usually have to use at least 3-4 different open source libraries to reach the last step of finding the right intervention. CausalNex aims to simplify the end-to-end process of causality and counterfactual analysis.
2 model used
2.1 structural equation model of notears
reference resources: DAG_GNN: a DAG structure learning architecture based on VAE
The open source library model refers to NOTEARS model: DAGs with NO TEARS: Continuous Optimization for Structure Learning
Several highlights in the paper:
- In this paper, the acyclic structure constraint of DAG learning is improved from the traditional combination constraint (discrete, acyclic structure increases in geometric multiples) to a smooth continuous constraint( h ( w ) h(w) h(w) )
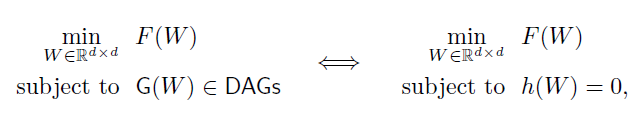

F(W) is the score function, and DAG_GNN is another upgrade model, which will be used in this
h
(
w
)
h(w)
h(w) upgrade;
- h ( w ) h(w) After h(w), X is modeled by the defined structural equation model (SEM), which focuses on finding a SEM that can minimize the loss of least squares (LS).
3 modeling case: NOTEARS structural equation model
3.1 data loading
Case source: Structure-Learning
import pandas as pd
data = pd.read_csv('student-por.csv', delimiter=';')
data.head(5)
# Data processing
drop_col = ['school','sex','age','Mjob', 'Fjob','reason','guardian']
data = data.drop(columns=drop_col)
data.head(5)
import numpy as np
struct_data = data.copy()
non_numeric_columns = list(struct_data.select_dtypes(exclude=[np.number]).columns)
# Data text - > numeric
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for col in non_numeric_columns:
struct_data[col] = le.fit_transform(struct_data[col])
struct_data.head(5)
After loading the data and performing some numerical operations on the data, the next step is modeling:
from causalnex.structure.notears import from_pandas sm = from_pandas(struct_data)

It can be seen here that each variable is interconnected because there is no expert assignment and no filtering edge operation, so it is displayed;
Alternatively, you can use remove_edges_below_threshold to set some thresholds for deletion:
sm.remove_edges_below_threshold(0.8)
viz = plot_structure(
sm,
graph_attributes={"scale": "0.5"},
all_node_attributes=NODE_STYLE.WEAK,
all_edge_attributes=EDGE_STYLE.WEAK,
)
Image(viz.draw(format='png'))
If the edge coefficient threshold is less than 0.8 is deleted, the current figure will appear:
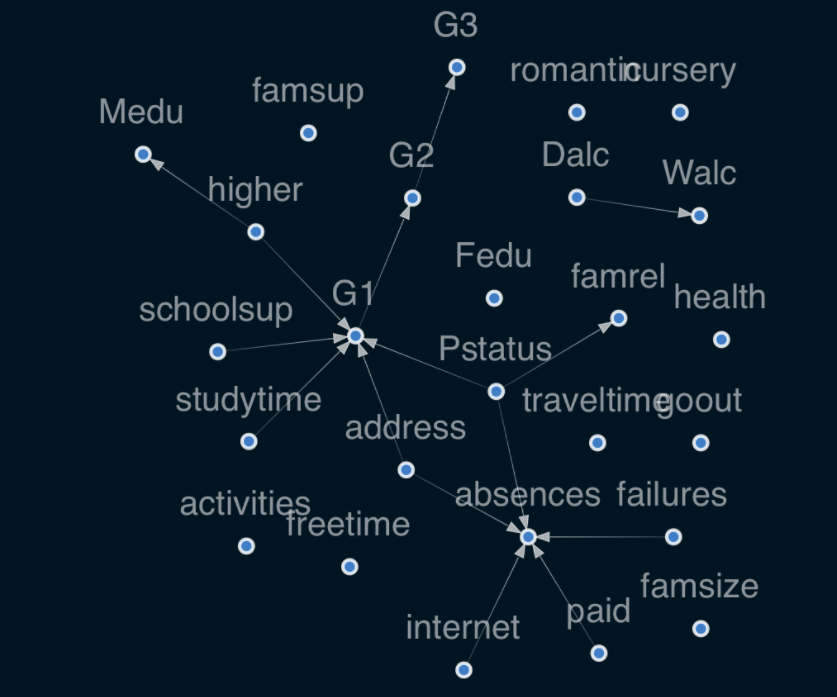
Interpretation of results:
- Family status P s t a t u s Pstatus Pstatus affects family relations f a m r e l famrel famrel - if parents are separated, the quality of family relations may deteriorate.
- network i n t e r n e t internet internet affects absenteeism a b s e n c e s absences absences - having an Internet at home may cause students to skip classes.
- Long learning time s t u d y t i m e studytime studytime on students' grades G 1 G1 G1 has a positive impact.
3.2 operations required for modeling & error pointing
3.2.1 error pointing solution 1: add constraints
Of course, there may also be a wrong direction:
Children's Higher Education
h
i
g
h
e
r
higher
higher will affect
M
e
d
u
Medu
Medu (mother's education) - this relationship is meaningless because students who want to pursue higher education will not affect their mother's education.
For this wrong direction, some constraints can be added during modeling:
sm = from_pandas(struct_data, tabu_edges=[("higher", "Medu")], w_threshold=0.8)
viz = plot_structure(
sm,
graph_attributes={"scale": "0.5"},
all_node_attributes=NODE_STYLE.WEAK,
all_edge_attributes=EDGE_STYLE.WEAK,
)
Image(viz.draw(format='png'))

3.2.2 error pointing solution 2: direct fine tuning results
Add / delete edge
sm.add_edge("failures", "G1")
sm.remove_edge("Pstatus", "G1")
sm.remove_edge("address", "G1")

3.3 retrieving the largest subgraph
We can see that there are two independent subgraphs in the visual diagram: dalc - > WALC and another large subgraph.
We can easily retrieve the largest subgraph by calling the StructureModel function get_largest_subgraph().
sm = sm.get_largest_subgraph()
viz = plot_structure(
sm,
graph_attributes={"scale": "0.5"},
all_node_attributes=NODE_STYLE.WEAK,
all_edge_attributes=EDGE_STYLE.WEAK,
)
Image(viz.draw(format='png'))
3.4 export structure
To export nx#u pydot using networkx:
import networkx as nx nx.drawing.nx_pydot.write_dot(sm, 'graph.dot')
3.5 fitting conditional distribution of Bayesian network: data discretization
When the edge structure information is confirmed, Bayesian network can be used to fit different characteristic conditional probability distributions.
Bayesian networks in CausalNex only support discrete distribution. Any continuous features, or features with a large number of categories, should be discretized before fitting Bayesian networks. Models containing variables with many possible values are usually not suitable and show poor performance.
For example, consider P(G2 | G1), where G1 and G2 have possible values of 0 to 20. Therefore, the discrete conditional probability distribution is 21 × 21 (441) possible combinations are specified - most of which we will be unlikely to observe.
CausalNex provides some auxiliary methods to simplify discretization. Let's start by reducing the number of categories in some classification features by combining similar values. We will classify digital features through discretization, and then give meaningful labels to buckets.
from causalnex.network import BayesianNetwork bn = BayesianNetwork(sm)
For example, in the learning time function, we put more than 2 hours (2 means 2 to 5 hours here, see https://archive.ics.uci.edu/ml/datasets/Student +The learning time of performance) is divided into long learning time and the rest into short learning time.
discretised_data = data.copy()
data_vals = {col: data[col].unique() for col in data.columns}
failures_map = {v: 'no-failure' if v == [0]
else 'have-failure' for v in data_vals['failures']}
studytime_map = {v: 'short-studytime' if v in [1,2]
else 'long-studytime' for v in data_vals['studytime']}
discretised_data["failures"] = discretised_data["failures"].map(failures_map)
discretised_data["studytime"] = discretised_data["studytime"].map(studytime_map)
Discretizing digital features
In order to make digital features classified, they must be discretized first.
CausalNex provides an auxiliary class causalnex.discretiser.Discretiser.
Supports a variety of discretization methods.
For our data, a fixed method will be applied to provide static values that define the bucket boundary.
For example, absenteeism will be divided into several parts < 1, 1-9, > = 10.
Each bucket will be marked as a zero based integer.
from causalnex.discretiser import Discretiser
discretised_data["absences"] = Discretiser(method="fixed",
numeric_split_points=[1, 10]).transform(discretised_data["absences"].values)
discretised_data["G1"] = Discretiser(method="fixed",
numeric_split_points=[10]).transform(discretised_data["G1"].values)
discretised_data["G2"] = Discretiser(method="fixed",
numeric_split_points=[10]).transform(discretised_data["G2"].values)
discretised_data["G3"] = Discretiser(method="fixed",
numeric_split_points=[10]).transform(discretised_data["G3"].values)
Create labels for numeric properties
To make discrete categories more readable, we can map category labels to something more meaningful, just like mapping category eigenvalues.
absences_map = {0: "No-absence", 1: "Low-absence", 2: "High-absence"}
G1_map = {0: "Fail", 1: "Pass"}
G2_map = {0: "Fail", 1: "Pass"}
G3_map = {0: "Fail", 1: "Pass"}
discretised_data["absences"] = discretised_data["absences"].map(absences_map)
discretised_data["G1"] = discretised_data["G1"].map(G1_map)
discretised_data["G2"] = discretised_data["G2"].map(G2_map)
discretised_data["G3"] = discretised_data["G3"].map(G3_map)
3.6 BN model training
Split the data, training set and test set:
# Split 90% train and 10% test from sklearn.model_selection import train_test_split train, test = train_test_split(discretised_data, train_size=0.9, test_size=0.1, random_state=7)
Using the previously learned structural model and discrete data, we can now fit the probability distribution of Bayesian network.
The first step is to specify all states that each node can take. This can be done by data or providing a node value dictionary.
We use the complete data set here to avoid the situation that the state in the test set does not exist in the training set.
For real applications, you might need to provide these states using the dictionary method.
# Model initialization bn = bn.fit_node_states(discretised_data) # Fitting conditional probability distribution bn = bn.fit_cpds(train, method="BayesianEstimator", bayes_prior="K2")
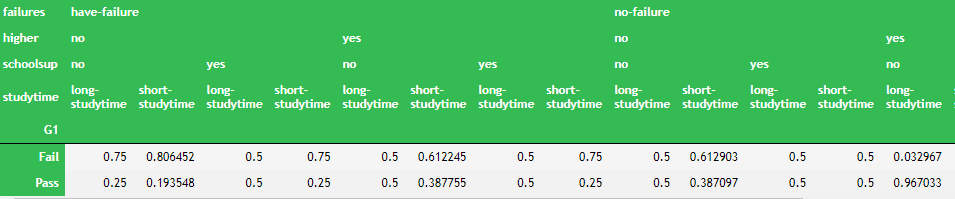
View the conditional probability distributions (CPDS) dictionary of BN model:
bn.cpds["G1"]
3.7 forecast given input data
The prediction method of Bayesian network enables us to use the learned Bayesian network to predict the data.
For example, we want to predict whether a student has passed the exam based on the input data.
Suppose we have an incoming student data, which is as follows:
discretised_data.loc[18, discretised_data.columns != 'G1']

predictions = bn.predict(discretised_data, "G1")
print(f"The prediction is '{predictions.loc[18, 'G1_prediction']}'")
Student No. 18, the study time here is expressed as short-study time, which feels impossible.
The prediction is 'Fail'
The prediction result here is also fail, which is reasonable.
3.8 model evaluation: classification report + ROC/AUC
3.8.1 Classification Report
from causalnex.evaluation import classification_report classification_report(bn, test, "G1")
Output results:
{'G1_Fail': {'precision': 0.7777777777777778,
'recall': 0.5833333333333334,
'f1-score': 0.6666666666666666,
'support': 12},
'G1_Pass': {'precision': 0.9107142857142857,
'recall': 0.9622641509433962,
'f1-score': 0.9357798165137615,
'support': 53},
'accuracy': 0.8923076923076924,
'macro avg': {'precision': 0.8442460317460317,
'recall': 0.7727987421383649,
'f1-score': 0.8012232415902141,
'support': 65},
'weighted avg': {'precision': 0.8861721611721611,
'recall': 0.8923076923076924,
'f1-score': 0.8860973888496825,
'support': 65}}
3.8.2 ROC/AUC
from causalnex.evaluation import roc_auc roc, auc = roc_auc(bn, test, "G1") print(auc)
3.9 Querying Marginals
3.9.1 basic query: baseline margins
If you want to query the probability values of some nodes, the previous model cannot be queried. Now you need to rebuild a query engine:
bn = bn.fit_cpds(discretised_data, method="BayesianEstimator", bayes_prior="K2")
from causalnex.inference import InferenceEngine
ie = InferenceEngine(bn)
marginals = ie.query()
marginals["G1"]
>>> {'Fail': 0.25260687281677224, 'Pass': 0.7473931271832277}
Get the conditional probability of each node. Here you can see the node G1. 0.252 is the number of fial and 0.747 is pass
The overall figures can be obtained for checking calculation, which is consistent
import numpy as np labels, counts = np.unique(discretised_data["G1"], return_counts=True) list(zip(labels, counts))
3.9.2 query criteria
We can also query the marginal likelihood of states in the network given some observations.
These observations can be made anywhere in the network, and their impact will spread through the nodes of interest.
Let's look at the possibility of G1 according to the study time.
marginals_short = ie.query({"studytime": "short-studytime"})
marginals_long = ie.query({"studytime": "long-studytime"})
print("Marginal G1 | Short Studtyime", marginals_short["G1"])
print("Marginal G1 | Long Studytime", marginals_long["G1"])
Output is:
Marginal G1 | Short Studtyime {'Fail': 0.2776556433482524, 'Pass': 0.7223443566517477}
Marginal G1 | Long Studytime {'Fail': 0.15504850337837614, 'Pass': 0.8449514966216239}
It can be seen here that 0.722 of G1 is passed if there is Short Studtyime status;
With Long Studytime status, 0.844 of G1 is passed
So relatively speaking, Long Studytime is easier to pass than short studytime.
3.10 [final / core link] Do Calculus - under confounding factor
We can apply intervention to any node in the data and update its distribution using the do operator. This can be thought of as asking our model "what if it's different".
For example, we can ask, what happens if 100% of students want to continue higher education:
print("marginal G1", ie.query()["G1"])
ie.do_intervention("higher",
{'yes': 1.0,
'no': 0.0})
print("updated marginal G1", ie.query()["G1"])
Conclusion:
marginal G1 {'Fail': 0.25260687281677224, 'Pass': 0.7473931271832277}
updated marginal G1 {'Fail': , 'Pass': 0.79?????}
It can be seen here that if 90% of people are willing to continue to receive higher education, the G1 score is 0.74;
If it rises to 100% willingness to accept, the average score of G1 will increase to 0.79
4 scikit learn integration interface
DAGRegressor / DAGClassifier is the regression / classification function of scikit learn
It looks more convenient to use here.
4.1 linear regression: Linear DAGRegressor
from sklearn.datasets import load_diabetes
import torch
torch.manual_seed(42)
data = load_diabetes()
X, y = data.data, data.target
names = data["feature_names"]
from sklearn.preprocessing import StandardScaler
ss = StandardScaler()
X = ss.fit_transform(X)
y = (y - y.mean()) / y.std()
from causalnex.structure import DAGRegressor
reg = DAGRegressor(
alpha=0.1,
beta=0.9,
hidden_layer_units=None,
dependent_target=True,
enforce_dag=True,
)
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
scores = cross_val_score(reg, X, y, cv=KFold(shuffle=True, random_state=42))
print(f'MEAN R2: {np.mean(scores).mean():.3f}')
X = pd.DataFrame(X, columns=names)
y = pd.Series(y, name="DPROG")
reg.fit(X, y)
print(pd.Series(reg.coef_, index=names))
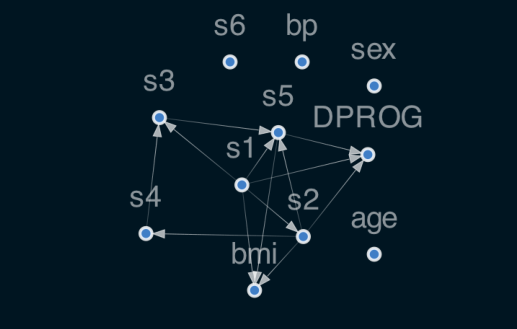
reg.plot_dag(enforce_dag=True)
Simultaneous output:
MEAN R2: 0.478 age 0.000000 sex 0.000000 bmi 0.304172 bp 0.000000 s1 0.000000 s2 0.000000 s3 0.000000 s4 0.000000 s5 0.277010 s6 0.000000 dtype: float64
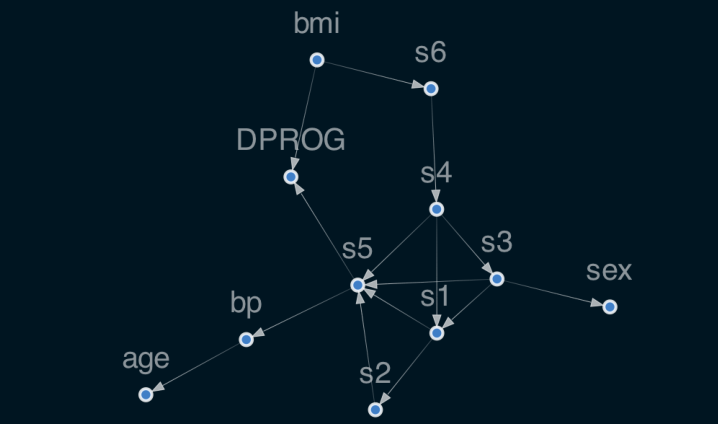
Here are some special model parameters,
- dependent_target: whether X - > y and Y - > x are the same. If it is true, it can point backwards. The default is true
- enforce_dag, whether to delete some edges when displaying the visualization diagram, but it will not be deleted when actually running the model. The default is on
4.2 nonlinear regression
In the process of data processing, it is important to standardize the data
from sklearn.datasets import load_diabetes
import torch
torch.manual_seed(42)
data = load_diabetes()
X, y = data.data, data.target
names = data["feature_names"]
from causalnex.structure import DAGRegressor
reg = DAGRegressor(
threshold=0.0,
alpha=0.0001,
beta=0.2,
hidden_layer_units=[2],
standardize=True,
enforce_dag=True,
)
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
scores = cross_val_score(reg, X, y, cv=KFold(shuffle=True, random_state=42))
print(f'MEAN R2: {np.mean(scores).mean():.3f}')
X = pd.DataFrame(X, columns=names)
y = pd.Series(y, name="DPROG")
reg.fit(X, y)
reg.plot_dag(enforce_dag=True)
The nonlinear layer contains s-type nonlinearity, which will be saturated by unscaled data.
In addition, the non scaled data means that the regularization parameters will not have the same impact on the weight of each feature.
Where, standardize = True, which means standardizing X/Y.
It also performs the inverse transformation of y on predict, similar to sklearn transformatedtargetregressor.
4.3 classification model: DAGClassifier
from sklearn.datasets import load_breast_cancer
data = load_breast_cancer()
X, y = data.data, data.target
names = data["feature_names"]
from causalnex.structure import DAGClassifier
clf = DAGClassifier(
alpha=0.1,
beta=0.9,
hidden_layer_units=[5],
fit_intercept=True,
standardize=True
)
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
scores = cross_val_score(clf, X, y, cv=KFold(shuffle=True, random_state=42))
print(f'MEAN Score: {np.mean(scores).mean():.3f}')
X = pd.DataFrame(X, columns=names)
y = pd.Series(y, name="NOT CANCER")
clf.fit(X, y)
for i in range(clf.coef_.shape[0]):
print(f"MEAN EFFECT DIRECTIONAL CLASS {i}:")
print(pd.Series(clf.coef_[i, :], index=names).sort_values(ascending=False))
clf.plot_dag(True)
Output:
MEAN Score: 0.975 MEAN EFFECT DIRECTIONAL CLASS 0: fractal dimension error 0.435006 texture error 0.253090 mean compactness 0.225626 symmetry error 0.112259 compactness error 0.094851 worst compactness 0.087924 mean fractal dimension 0.034403 concavity error 0.030805 mean texture 0.024233 mean symmetry -0.000012 mean perimeter -0.000603 mean area -0.003670 mean radius -0.005750 mean smoothness -0.007812 perimeter error -0.021268 concave points error -0.088604 smoothness error -0.173150 worst smoothness -0.174532 worst fractal dimension -0.193018 worst perimeter -0.241930 worst concavity -0.251617 worst symmetry -0.253782 worst concave points -0.255410 mean concavity -0.317477 mean concave points -0.322275 worst area -0.427581 worst radius -0.448448 radius error -0.489342 area error -0.518434 worst texture -0.614594 dtype: float64
