Project background
A recent report by the food and Agriculture Organization of the United Nations shows that more than one third of the natural losses of agricultural production each year are caused by agricultural diseases and pests. Many agricultural diseases and pests need to be considered. The traditional methods that rely on laboratory observation and experiment are easy to lead to wrong diagnosis. In order to accelerate the transformation of agricultural development mode, the Ministry of Agriculture organized the integration of specialized unified control and green prevention and control of crop diseases and pests, and gradually realized the large-scale implementation and standardized operation of whole process green prevention and control of crop diseases and pests. Integration and promotion can effectively improve the organization and scientific level of disease and pest control. It is not only an important content of comprehensive pest control and pesticide reduction and control, but also a major measure to change the mode of agricultural development and improve quality and efficiency. While ensuring the control effect, the quality of agricultural products met the national food safety standards, and the ecological environment and biodiversity were improved.

Project purpose
As long as crop diseases and pests occur, it will not only have a very serious impact on people's life and environment, but also have an adverse impact on Farmers' income.
In the vast rural areas, most farmers prefer traditional pesticides for pest control:
-
Poor awareness of pest control;
-
The application of traditional pesticides does not accord with the actual situation, such as unreasonable dosage, unreasonable cycle and so on. It not only has no effective prevention and control, but also causes great damage to crops, but also has the problem of serious environmental pollution. Finally, the effect of crop disease control is very poor, so that crops can not grow normally. In the end, the difficulty of manual prevention and control increases, the cost increases, and the consequences are serious.
-
In order to accelerate the transformation of agricultural development mode, the Ministry of Agriculture organized the integration of specialized unified control of crop diseases and insect pests and green prevention and control, and gradually realized the large-scale implementation and standardized operation of whole process green prevention and control of crop diseases and insect pests. Integration and promotion can effectively improve the organization and scientific level of disease and pest control. It is not only an important content of comprehensive pest control and pesticide reduction and control, but also a major measure to change the mode of agricultural development and improve quality and efficiency. While ensuring the control effect, the quality of agricultural products met the national food safety standards, and the ecological environment and biodiversity were improved.
Apply in-depth learning to accurately identify the diseases of crops and recommend appropriate control measures, so as to create a "plant doctor" who can see a doctor for crops, which plays the effect and standard of comprehensive control to a certain extent, so as to better eliminate crop diseases, promote the healthy growth of crops and ensure the yield of crops.
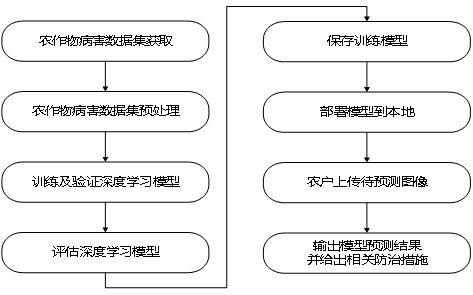
Technology Roadmap
System architecture diagram
Project innovation
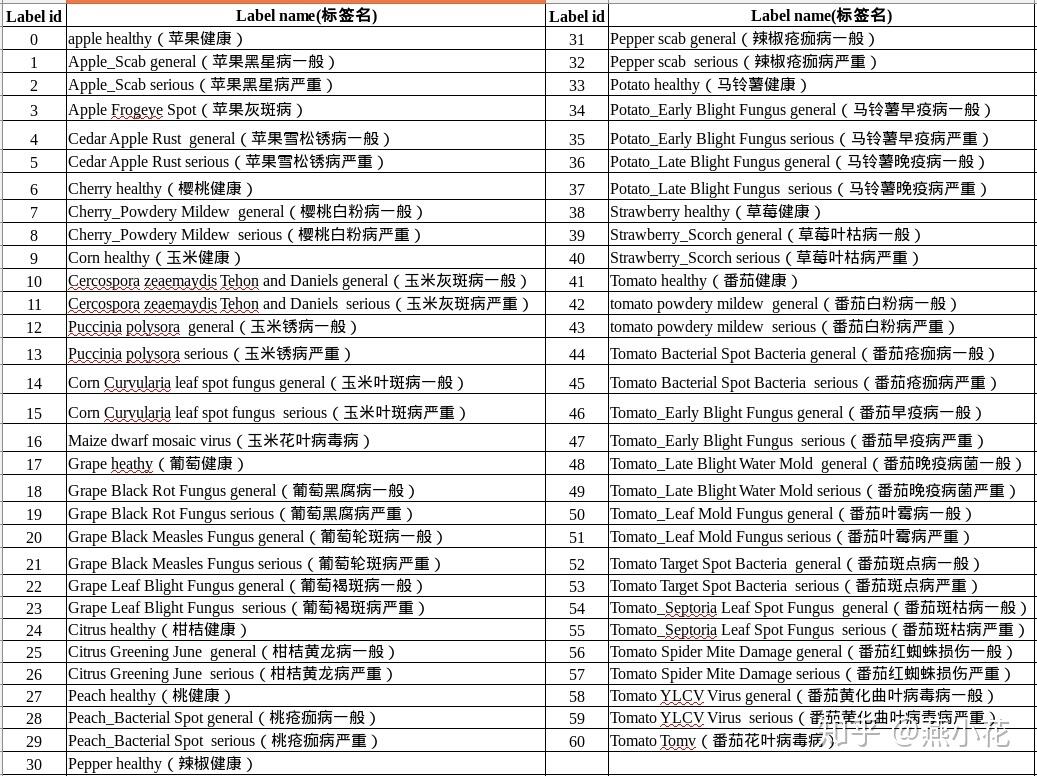
Based on the characteristics of the AI Challenger crop leaf image data set, which contains 27 diseases of 10 plants, a total of 61 classifications (divided by "species disease degree"), and combined with the data collected on the spot, this work will mainly consider the improvement from the following aspects:
- ① Increase certain crop types and subdivision granularity
Most of the diseases and pests that can be identified are too late to be identified. For example, the early symptoms of Strawberry Powdery mildew are very insignificant (very thin white villous spots), which can be seen only when a large number of white powdery fungi are full of leaves (or fruits). - ② Optimize the model, pay attention to the prediction accuracy and increase considerations
There are some diseases and insect pests that cannot see the disease / insect source at the root before the harm occurs, such as nematodes in plant roots and aphids hidden behind leaves. The monitoring is not limited to image analysis and recognition. - ③ Retrospective prediction of disease factors
At present, the system in the world only has a simple identification function and lacks the analysis of the causes of diseases and pests. This project will improve and improve this direction
data set
Based on the characteristics of AI Challenger crop leaf image data set, this study contains 27 diseases of 10 plants (apple, cherry, grape, orange, peach, strawberry, tomato, pepper, corn and potato) (24 diseases are divided into general and serious degrees), with a total of 61 classifications (according to "species disease degree"), and the total number of training images is 31718, The total number of test images is 4540. Each picture contains a leaf of a crop, which occupies the main position of the picture.
network model
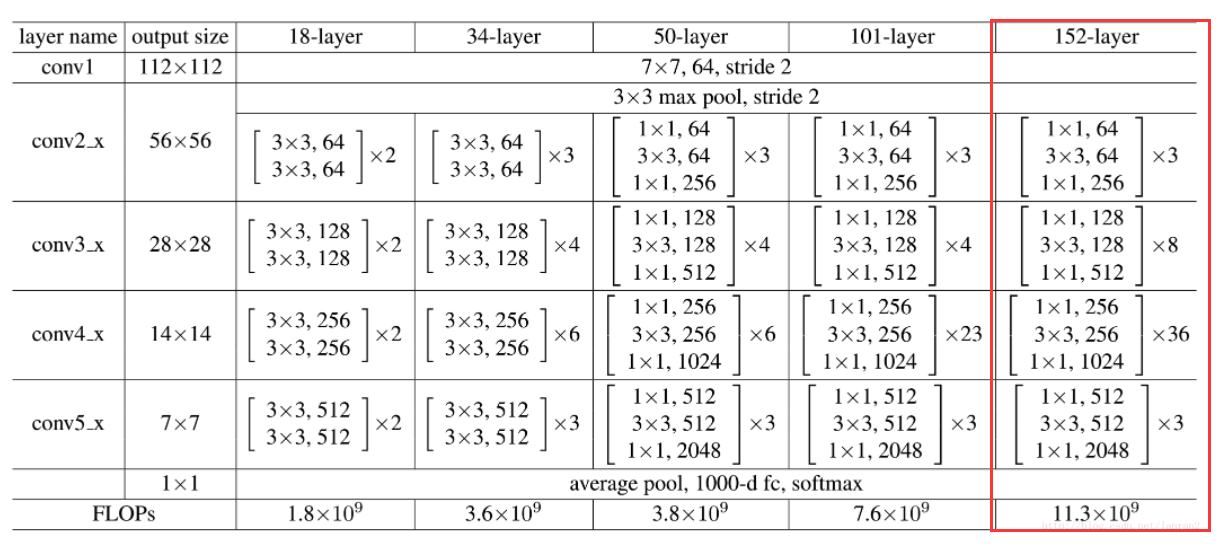
ResNet solves the problems of loss and loss in information transmission of traditional convolution layer or full connection layer by changing the learning goal, that is, it no longer learns the complete output F(x), but learns the residual H(x) − X. The integrity of the information is protected to some extent by directly bypassing the information from the input to the output. At the same time, because the goal of learning is residual, the difficulty of learning is simplified.
Decompress dataset
#!unzip /home/aistudio/data/data101323/data.zip
Data preprocessing
import paddle
import paddle.nn.functional as F
import numpy as np
import cv2
import json
import math
import random
import os
from paddle.io import Dataset # Import Datasrt Library
filename = "AgriculturalDisease_trainingset/AgriculturalDisease_train_annotations.json"
f_open = open(filename)
fileJson = json.load(f_open)
train_data = []
for i in range(len(fileJson)):
img1=cv2.imread("AgriculturalDisease_trainingset/images/"+fileJson[i]['image_id'])
img2=cv2.resize(img1, (128,128), interpolation=cv2.INTER_AREA)/255
r=[]
g=[]
b=[]
r.append(img2[:, :, 0])
g.append(img2[:, :, 1])
b.append(img2[:, :, 2])
one_data = np.concatenate((r,g,b),axis=0)
one_data = paddle.to_tensor(one_data,dtype="float32")
train_data.append([one_data,fileJson[i]['disease_class']])
filename = "AgriculturalDisease_validationset/AgriculturalDisease_validation_annotations.json"
f_open = open(filename)
fileJson1 = json.load(f_open)
test_data = []
for i in range(len(fileJson1)):
img1=cv2.imread("AgriculturalDisease_validationset/images/"+fileJson1[i]['image_id'])
img2=cv2.resize(img1, (128,128), interpolation=cv2.INTER_AREA)/255
r=[]
g=[]
b=[]
r.append(img2[:, :, 0])
g.append(img2[:, :, 1])
b.append(img2[:, :, 2])
one_data = np.concatenate((r,g,b),axis=0)
one_data = paddle.to_tensor(one_data,dtype="float32")
test_data.append([one_data,fileJson1[i]['disease_class']])
/opt/conda/envs/python35-paddle120-env/lib/python3.7/site-packages/paddle/fluid/layers/utils.py:26: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information. Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations def convert_to_list(value, n, name, dtype=np.int):
data fetch
from paddle.static import InputSpec
import paddle.nn.functional as F
print("-------end readData--------")
class MyDataset(Dataset):
"""
Step 1: inherit paddle.io.Dataset class
"""
def __init__(self, mode='train'):
"""
Step 2: implement the constructor, define the data reading method, and divide the training and test data sets
"""
super(MyDataset, self).__init__()
if mode == 'train':
self.data = train_data
else:
self.data = test_data
def __getitem__(self, index):
"""
Step 3: Implement__getitem__Methods, defining and specifying index How to obtain data and return a single piece of data (training data, corresponding label)
"""
data = self.data[index][0]
label = self.data[index][1]
return data, label
def __len__(self):
"""
Step 4: Implement__len__Method to return the total number of data sets
"""
return len(self.data)
# s_tra_data,s_tra_label = split_data(train_data,train_label,batch_size=32)
# s_tes_data,s_tes_label = split_data(test_data,test_label,batch_size=32)
#data fetch
train_loader = paddle.io.DataLoader(MyDataset("train"), batch_size=16, shuffle=True)
test_loader = paddle.io.DataLoader(MyDataset("test"), batch_size=16, shuffle=True)
-------end readData--------
Define training process
epoch_num = 20 #Number of training rounds
batch_size = 16
learning_rate = 0.0001 #Learning rate
val_acc_history = []
val_loss_history = []
def train(model):
print('start training ... ')
# turn into training mode
model.train()
opt = paddle.optimizer.Adam(learning_rate=learning_rate,
parameters=model.parameters())
for epoch in range(epoch_num):
acc_train = []
for batch_id, data in enumerate(train_loader()):
x_data = data[0]
y_data = paddle.to_tensor(data[1],dtype="int64")
y_data = paddle.unsqueeze(y_data, 1)
logits = model(x_data)
loss = F.cross_entropy(logits, y_data)
acc = paddle.metric.accuracy(logits, y_data)
acc_train.append(acc.numpy())
if batch_id % 200 == 0 and batch_id != 0:
print("epoch: {}, batch_id: {}, loss is: {}".format(epoch, batch_id, loss.numpy()))
avg_acc = np.mean(acc_train)
print("[train] accuracy: {}".format(avg_acc))
loss.backward()
opt.step()
opt.clear_grad()
# evaluate model after one epoch
model.eval()
accuracies = []
losses = []
for batch_id, data in enumerate(test_loader()):
x_data = data[0]
y_data = paddle.to_tensor(data[1],dtype="int64")
y_data = paddle.unsqueeze(y_data, 1)
logits = model(x_data)
loss = F.cross_entropy(logits, y_data)
acc = paddle.metric.accuracy(logits, y_data)
accuracies.append(acc.numpy())
losses.append(loss.numpy())
avg_acc, avg_loss = np.mean(accuracies), np.mean(losses)
print("[test] accuracy/loss: {}/{}".format(avg_acc, avg_loss))
val_acc_history.append(avg_acc)
val_loss_history.append(avg_loss)
model.train()
Start training
model = paddle.vision.models.resnet152(pretrained=True,num_classes=61) train(model) path = "save_model" model.train()
Start training
model = paddle.vision.models.resnet152(pretrained=True,num_classes=61) train(model) path = "save_model" paddle.jit.save(model, path,input_spec=[InputSpec(shape=[16,3,128,128], dtype='float32')])
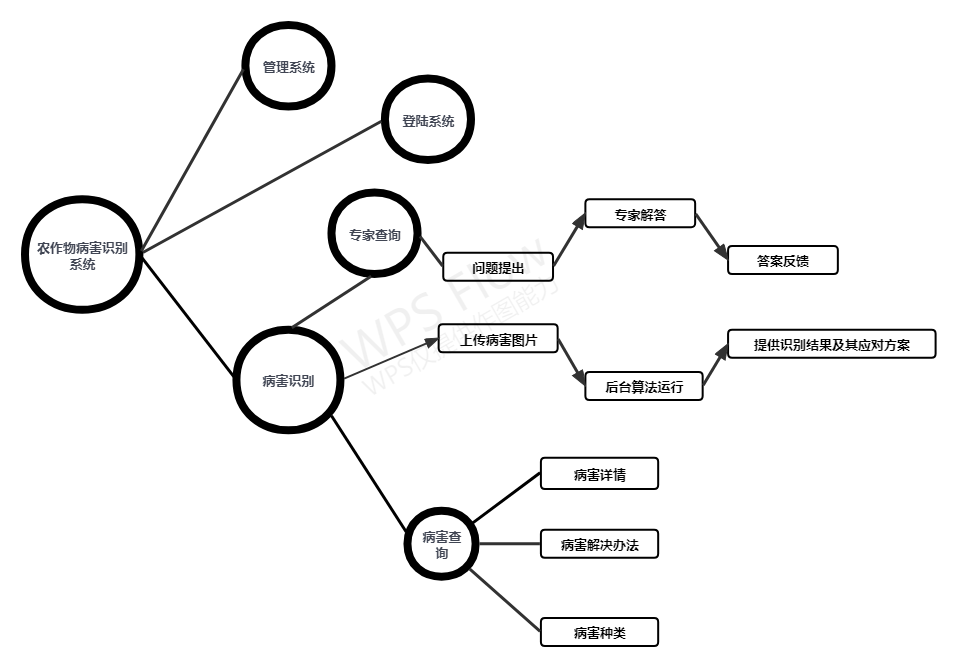
Front end implementation of works
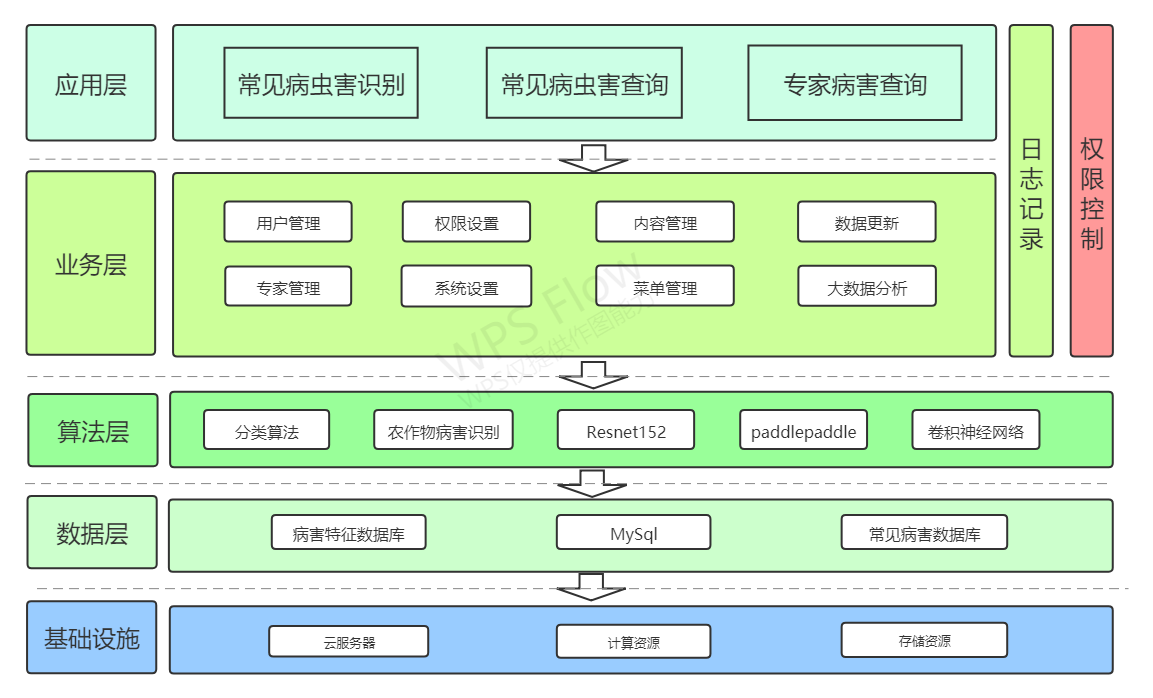
Front end function structure diagram
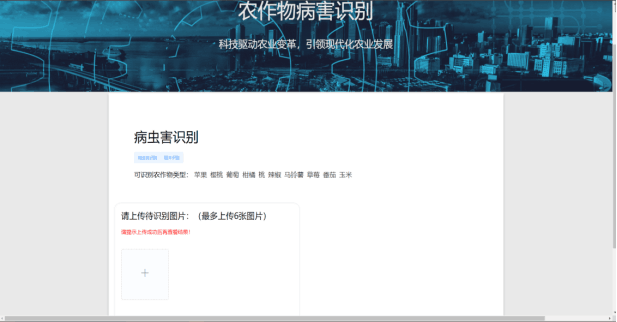
System main interface

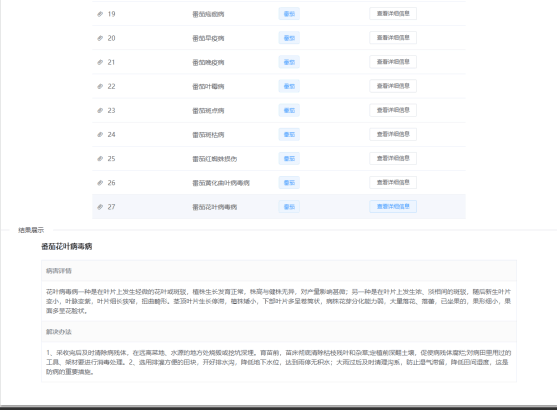
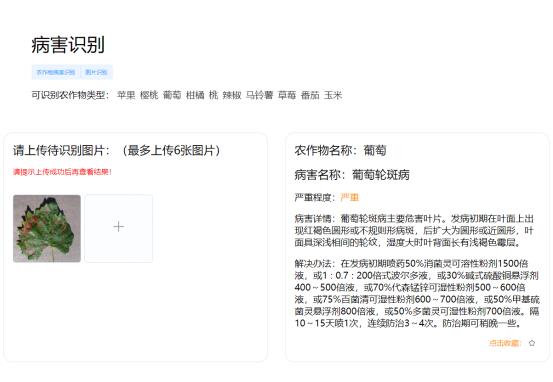
Pest query interface
Disease identification results
Common disease query