K-fold cross validation:
sklearn.model_selection.KFold(n_splits=3, shuffle=False, random_state=None)
Idea: the training / test data set is divided into n ﹣ splits mutually exclusive subsets, one of which is used as the verification set each time, and the remaining N ﹣ splits-1 is used as the training set for n ﹣ splits training and testing, and N ﹣ splits results are obtained
Note: for unequal data sets, the first n ﹣ samples% n ﹣ splits subset has n ﹣ samples / / N ﹣ splits + 1 sample, and the rest subset only has n ﹣ samples / / N ﹣ splits sample
Parameter Description:
N ﹣ splits: indicates how many equal parts are divided
Shuffle: shuffle or not during each division
① If false, the effect is the same as that of random state, which is the same as that of integer
② If it is True, the result of each partition is different, which means that after shuffling, random sampling
Random state: random seed number
Properties:
① Get ﹣ n ﹣ splits (x = none, y = none, groups = none): get the value of parameter n ﹣ splits
② split(X, y=None, groups=None): divides the data set into training set and test set, and returns the index generator
Through a chestnut that can not be divided equally, set different parameter values and observe the results
① Set shuffle=False, run twice, and the results are the same
from mlxtend.classifier import StackingClassifier sclf = StackingClassifier(classifiers=[lgb], meta_classifier=xgb_model) sclf_score=sclf.fit(train,target) test_predict=sclf.predict(test) from sklearn.metrics import r2_score def online_score(pred): print("Maximum predicted result:{},Minimum forecast result:{}".format(pred.max(),pred.min())) # a list conmbine1 = pd.read_csv(r'C:\Users\lxc\Desktop\featurecup\sub_b_919.csv',engine = "python",header=None) score1 = r2_score(pred, conmbine1) print("Compare 919 points:{}".format(score1)) score = online_score(test_predict) //Maximum predicted result:19051.067151217972,Minimum forecast result:1199.97082591554 //Contrast919Fraction:0.981891385946527
Stacking
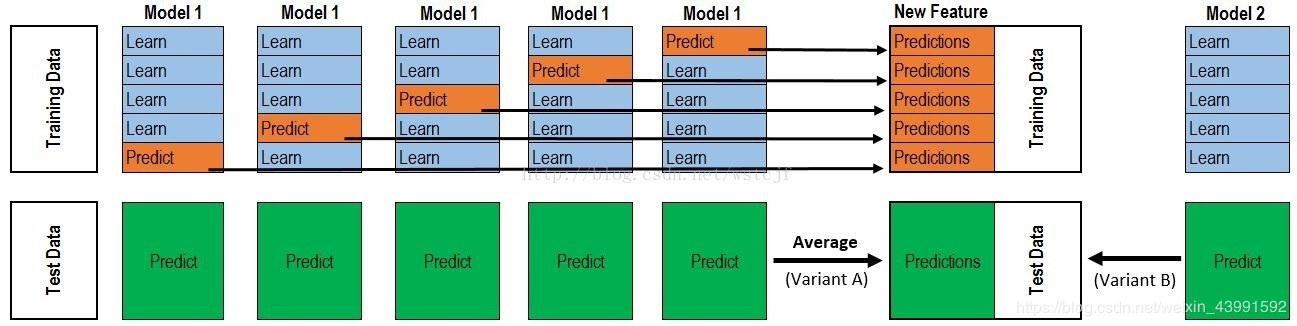
#!pip install mlxtend import warnings warnings.filterwarnings('ignore') import itertools import numpy as np import seaborn as sns import matplotlib.pyplot as plt import matplotlib.gridspec as gridspec from sklearn import datasets from sklearn.linear_model import LogisticRegression from sklearn.neighbors import KNeighborsClassifier from sklearn.naive_bayes import GaussianNB from sklearn.ensemble import RandomForestClassifier from mlxtend.classifier import StackingClassifier from sklearn.model_selection import cross_val_score, train_test_split from mlxtend.plotting import plot_learning_curves from mlxtend.plotting import plot_decision_regions
# Take the iris data set of python as an example iris = datasets.load_iris() X, y = iris.data[:, 1:3], iris.target clf1 = KNeighborsClassifier(n_neighbors=1) clf2 = RandomForestClassifier(random_state=1) clf3 = GaussianNB() lr = LogisticRegression() sclf = StackingClassifier(classifiers=[clf1, clf2, clf3], meta_classifier=lr) label = ['KNN', 'Random Forest', 'Naive Bayes', 'Stacking Classifier'] clf_list = [clf1, clf2, clf3, sclf] fig = plt.figure(figsize=(10,8)) gs = gridspec.GridSpec(2, 2) grid = itertools.product([0,1],repeat=2) clf_cv_mean = [] clf_cv_std = [] for clf, label, grd in zip(clf_list, label, grid): scores = cross_val_score(clf, X, y, cv=3, scoring='accuracy') print("Accuracy: %.2f (+/- %.2f) [%s]" %(scores.mean(), scores.std(), label)) clf_cv_mean.append(scores.mean()) clf_cv_std.append(scores.std()) clf.fit(X, y) ax = plt.subplot(gs[grd[0], grd[1]]) fig = plot_decision_regions(X=X, y=y, clf=clf) plt.title(label) plt.show()
Blending
Blending method It's another way of integrating models that is different from bagging and boosting.
In the case of multiple weak learners, how to combine the prediction values of these weak learners to get better prediction value is what Blending does.
def blend(train,test,target): '''5 fracture''' # n_flods = 5 # skf = list(StratifiedKFold(y, n_folds=n_flods)) '''The segmentation training data set is d1,d2 The two part''' X_d1, X_d2, y_d1, y_d2 = train_test_split(train, target, test_size=0.5, random_state=914) train_ = np.zeros((X_d2.shape[0],len(clfs*3))) test_ = np.zeros((test.shape[0],len(clfs*3))) for j,clf in enumerate(clfs): '''Training each single model in turn''' # print(j, clf) '''The first part is used as the prediction, and the second part is used to train the model, and the predicted output is obtained as the new feature of the second part.''' # X_train, y_train, X_test, y_test = X[train], y[train], X[test], y[test] X_d1fillna=X_d1.fillna(0) X_d2fillna = X_d2.fillna(0) X_predictfillna= test.fillna(0) clf.fit(X_d1fillna,y_d1) y_submission = clf.predict(X_d2fillna) y_test_submission = clf.predict(X_predictfillna) train_[:,j*3] = y_submission*y_submission '''For test sets, use this directly k The predicted values of models are used as new features.''' test_[:, j*3] = y_test_submission*y_test_submission train_[:, j+1] =(y_submission - y_submission.min()) /(y_submission.max() - y_submission.min()) '''For test sets, use this directly k The predicted values of models are used as new features.''' y_test_submission = (y_test_submission - y_test_submission.min()) / \ (y_test_submission.max() - y_test_submission.min()) test_[:, j+1] = y_test_submission train_[:, j+2] = np.log(y_submission) '''For test sets, use this directly k The predicted values of models are used as new features.''' y_test_submission =np.log(y_test_submission) test_[:, j+2] = y_test_submission # print("val auc Score: %f" % r2_score(y_predict, dataset_d2[:, j])) print('Completed',j) train_.to_csv('./input/train_blending.csv', index=False) test_.to_csv('./input/test_blending.csv', index=False)

