0 declaration
The main content of this article comes from video ' [2020 complete set of machine learning] the complete version of sklearn of vegetables, the most complete complete complete set of machine learning sklearn worth 4999 yuan, quickly collect bilibili bili 'courseware from“ https://pan.baidu.com/s/1Xl4o0PMA5ysUILeCKvm_2w , extraction code: a967 ".
In addition to referring to the above videos and other materials, ' [Feature Engineering] disgusting work - in depth understanding of Feature Engineering _wx:wu805686220-CSDN blog 'it's also very helpful to me.
This article is a learning note, not an article to help newcomers get started. Its content is mainly aimed at my weak links. It is not comprehensive and may not be applicable to everyone.
1 data preprocessing
1.1 dimensionless data
Dimensionless enables data of different specifications to be converted to the same specification. The common dimensionless methods are standardization and normalization. In the algorithms with gradient and matrix as the core, such as logistic regression, support vector machine, neural network, dimensionless can speed up the solution speed; In distance models, such as k-nearest neighbors and K-Means clustering, dimensionless can improve the accuracy of the model and avoid the impact of a feature with a particularly large value range on distance calculation. (a special case is the integration algorithm of decision tree and tree. The decision tree does not need dimensionless. The decision tree can handle any data well.)
1.1. 1 normalization or min max scaling
After normalization, the data will obey the normal distribution. The normalization formula is as follows:
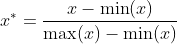
Where min(x) represents the minimum value of each column of data, max(x) is the maximum value of each column of data, and X * is the normalization result
'''Normalize each column (feature)''' from sklearn.preprocessing import MinMaxScaler # Import normalization module # feature_range controls the range of compressed data. The default is [0,1] scaler = MinMaxScaler(feature_range=[low,up]) # instantiation result_ = scaler.fit_transform(data) # Normalized data results data = scaler.inverse_transform(result) # Restore data to before normalization
1.1. 2 Standardization or Z-score normalization
After standardization, the data will obey the standard normal distribution, with the mean value of 0 and the variance of 1
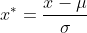
Where miu is the mean value of each column, sigma is the standard deviation of each column, and x * is the standardization result.
'''Standardize each column of data''' from sklearn.preprocessing import StandardScaler # Import standardization Toolkit scaler = StandardScaler() # instantiation x_std = scaler.fit_transform(data) # Complete standardization x = scaler.inverse_transform(x_std) # Restore data to before standardization
1.1. 3 selection of standardization and normalization
In most machine learning algorithms, StandardScaler is selected for feature scaling because MinMaxScaler is very sensitive to outliers. In PCA, clustering, logistic regression, support vector machine and neural network, StandardScaler is often the best choice; MinMaxScaler is widely used when it does not involve distance measurement, gradient, covariance calculation and data needs to be compressed to a specific interval. For example, when quantifying pixel intensity in digital image processing, MinMaxScaler will be used to compress data into [0,1] interval. In practical use, you can try both.
1.2 missing value handling
In addition to deleting features or samples with missing values, you can also fill them. The general filling methods include constant filling, mean filling, mode filling, uplink and downlink data filling and mode filling
1.2. 1 fill in sklearn
from sklearn.impute import SimpleImputer # Import population Kit Age = data.loc[:,"Age"].values.reshape(-1,1) # The characteristic matrix in sklearn must be two-dimensional values gets a one-dimensional array imp_mean = SimpleImputer() # Instantiation, default mean filling imp_median = SimpleImputer(strategy="median") # Fill with median imp_0 = SimpleImputer(strategy="constant",fill_value=0) # Fill with 0 imp_most_frequent = SimpleImputer(strategy="most_frequent") # Fill with mode imp_mean = imp_mean.fit_transform(Age) # fit_transform completes the retrieval result in one step imp_median = imp_median.fit_transform(Age) imp_0 = imp_0.fit_transform(Age) imp_most_frequent = imp_most_frequent.fit_transform(Age)
1.2. 2. Filling in pandas
import pandas as pd
data['Gray scale division'] = data['Gray scale division'].fillna('-99') # Fill with fixed values
data['Gray scale division'] = data['Gray scale division'].fillna(data['Gray scale division'].mean())) # Fill with mean
data['Gray scale division'] = data['Gray scale division'].fillna(data['Gray scale division'].mode())) # Fill with mode
data['Gray scale division'] = data['Gray scale division'].fillna(method='pad') # Populate with previous data
data['Gray scale division'] = data['Gray scale division'].fillna(method='bfill') # Fill with the last data
data['Gray scale division'] = data['Gray scale division'].interpolate() # Fill with interpolation1.2. 3 fill with algorithm
You can also use random forest, KNN and other algorithms to fill in the missing data. The filling effect is generally better than filling directly with mean, mode, etc.
1.3} feature conversion
1.3. 1 discrete data characteristics
In machine learning, most algorithms, such as logistic regression, support vector machine, SVM, k-nearest neighbor algorithm, etc., can only process numerical data, not text. However, in reality, many labels and features are not expressed in numbers when the data is collected. In order to adapt the data to the algorithm and Library, the data must be encoded, Convert text data to numeric data.
If variables are completely independent and cannot be compared or calculated, such variables are called nominal variables, such as cabin seat number. Nominal variables cannot be converted into numerical values by using OrdinalEncoder and LabelEncoder, but OneHotEncoder should be used, because OneHotEncoder can tell the algorithm that these are independent data and cannot be compared and calculated;
from sklearn.preprocessing import OneHotEncoder X = data.iloc[:,1] result = OneHotEncoder(categories='auto').fit_transform(X).toarray() name = result.get_feature_names() # Get the attribute corresponding to each code, and each column code represents an attribute X = pd.DataFrame(enc.inverse_transform(result)) # Restore to data before encoding
If there is a concept of size between variables, but no operation can be performed, such variables are called ordered variables, such as education. Ordered variables can be converted into numerical type by using OrdinalEncoder and LabelEncoder;
If there is a concept of size between variables, operations can also be carried out. This variable is called a distant variable. Such as height and weight. Spaced variables can be converted to numeric values using OrdinalEncoder and LabelEncoder.
from sklearn.preprocessing import LabelEncoder # This is a special class that converts labels to numeric types y = data.iloc[:,-1] # The label is to be entered, not the characteristic matrix, so one dimension is allowed le = LabelEncoder() # instantiation label = le.fit_transform(y) le.classes_ # Properties classes_ See how many classes are in the label data.iloc[:,-1] = label # Let the tag be equal to the result of our run, and the whole coding is over y = le.inverse_transform(label) #Use inverse_transform can be reversed back before encoding # Here is a more concise form data.iloc[:,-1] = LabelEncoder().fit_transform(data.iloc[:,-1]) from sklearn.preprocessing import OrdinalEncoder # Feature specific, which can convert classification features into classification values OrdinalEncoder().fit(data_.iloc[:,1:-1]).categories_ # See how many classes are in the label data_.iloc[:,1:-1] = OrdinalEncoder().fit_transform(data_.iloc[:,1:-1])
1.3. 2 continuity data characteristics
1.3. 2.1 binarization
The binarization of features is to output numerical data to boolean type. Its core is to set a threshold. When the number of samples is greater than the threshold, the output is 1, and when it is less than or equal to the threshold, the output is 0.
from sklearn.preprocessing import Binarizer # Import modules required for binarization X = data_2.iloc[:,0].values.reshape(-1,1) #Class is feature specific, so one-dimensional arrays cannot be used transformer = Binarizer(threshold=30).fit_transform(X)
1.3. 2.2 box sorting (numerical type is converted to category type)
The continuous variables are divided into categories of classified variables. The continuous variables can be sorted, boxed and coded in order. It mainly uses preprocessing Kbinsdiscretizer class. This class mainly contains three parameters:
| n_bins | The number of boxes in each feature, which is 5 by default, will be applied to all imported features at one time |
| encode | The feature encoding method is "onehot" by default "ordinal": each box of each feature is encoded as an integer, and each column is a feature. The matrix of boxes with different integer codes under each feature is returned; "Onehot dense": make a dummy variable, and then return a dense array |
| strategy | It is used to define the box width. The default is "quantile"; "uniform": indicates equal width boxes, that is, the difference between the maximum values of each box in each feature is (feature. max() - feature min())/(n_bins); "quantile": indicates equipotential bin division, that is, the number of samples in each bin in each feature is the same; "kmeans": indicates that the cluster is divided into boxes according to clustering, and the distance from the value in each box to the nearest cluster center of one-dimensional k-means clustering is the same |
from sklearn.preprocessing import KBinsDiscretizer X = data.iloc[:,0].values.reshape(-1,1) est = KBinsDiscretizer(n_bins=3, encode='ordinal', strategy='uniform') est.fit_transform(X) # View the boxes divided after conversion: it becomes three boxes in a column set(est.fit_transform(X).ravel()) est = KBinsDiscretizer(n_bins=3, encode='onehot', strategy='uniform') # View the sub box after conversion: it has become a dummy variable est.fit_transform(X).toarray()
2 feature selection
Not all features are equal. Selecting a small number of useful features from a large number of features is the so-called feature selection. Feature selection mainly includes filtering method, embedding method and Wrapper method.
2.1 filtration method
Filtering methods are usually used as preprocessing steps, and feature selection is completely independent of any machine learning algorithm. It selects features according to the scores in various statistical tests and various indicators of correlation. The main object of filtering method is the algorithm that needs to traverse features or upgrade dimensions, and the main purpose of filtering method is to help the algorithm reduce the computational cost on the premise of maintaining the performance of the algorithm. Nearest neighbor algorithm KNN, single decision tree, support vector machine, SVM, neural network and regression algorithm all need to traverse features or raise dimensions for operation, so they have a large amount of operation and take a long time. Therefore, feature selection such as variance filtering is particularly important for them. The random forest randomly selects features for branching, which is very fast, so the effect of feature selection is mediocre for it - no matter how the filtering method reduces the number of features, the random forest will only select a fixed number of features for modeling. The filtering method includes variance filtering and correlation filtering.
2.1. 1 variance filtering
Variance filtering filters the class of features through the variance of the feature itself. For example, if the variance of a feature itself is very small, it means that the samples have basically no difference in this feature. Maybe most values in the feature are the same, or even the values of the whole feature are the same, then this feature has no effect on sample differentiation.
from sklearn.feature_selection import VarianceThreshold selector = VarianceThreshold() # Instantiation. Fill in the value of variance in parentheses. If no parameter is filled, the variance is 0 by default X_var0 = selector.fit_transform(X) # Obtain the new feature matrix after deleting the unqualified feature
2.1. 2 selection of variance threshold
To select the optimal variance threshold, you can draw a learning curve to find the best point of the model. But in reality, it is often not done, because it will take a lot of time. Only variance filtering with a threshold of 0 or a small threshold will be used to eliminate some obviously unused features, and then a better feature selection method will be selected to continue to reduce the number of features.
2.1. 3. A thought
Why is the filtering method ineffective for random forest, but effective for tree model?
In terms of algorithm principle, the traditional decision tree needs to traverse all features and branch after calculating the impure, while the random forest randomly selects features for calculation and branching. Therefore, the operation of random forest is faster, and the filtering method is useless for random forest, but useful for decision tree.
In sklearn, both decision tree and random forest branch by randomly selecting features, but the number of features randomly extracted by decision tree in the modeling process is far more than that randomly extracted by each tree in random forest. Therefore, the filtering method is useless for the random forest, but it is useful for the decision tree. Therefore, in sklearn, each tree in the random forest is much simpler than a single decision tree, and the calculation of the random forest under high-dimensional data is much faster than the decision tree.
2.2 correlation filtering
After variance selection, the next problem should be considered: the correlation between tags and features. It is necessary to select meaningful features related to the label, because such features can provide a lot of information. If the feature has nothing to do with the label, it will only waste computing memory and may bring noise to the model. In sklearn, there are three common methods to evaluate the correlation between features and labels: Chi square test, F test and mutual information test.
2.2. 1 chi square test
Chi square test is used to test the correlation between qualitative independent variables and qualitative dependent variables, It is specially used for correlation filtering of discrete labels (i.e. classification problems). The chi square test class feature_selection.chi2 calculates the chi square statistics between each non negative feature and label, and ranks the features according to the chi square statistics from high to low. Combined with feature_selection.SelectKBest, it can enter the "scoring standard" "To select the class with the highest score of the first K features, so as to remove the features that are most likely independent of the label and irrelevant to the classification purpose.
from sklearn.feature_selection import SelectKBest # Pick features from sklearn.feature_selection import chi2 # Perform chi square test X_fschi = SelectKBest(chi2, k=300).fit_transform(X_fsvar, y) # k is the feature to be selected # Chi 2 perform chi square test # X_fsvar is the feature to be verified and y is the tag
The difficulty of chi square test lies in the selection of parameter K. the conventional method is to draw the learning curve, but the time of running this curve is very long, and it is generally not considered in large estimation data. Another efficient method is p-value screening. The essence of chi square test is to speculate the difference between the two groups of data, and its original hypothesis is that "the two groups of data are independent of each other". Chi square test returns two statistics: Chi square value and P value. Among them, chi square value is difficult to define the effective range. Generally, P is equal to 0.01 or 0.05 as the significance level, that is, the boundary of P value judgment. When p is greater than 0.01 or 0.05, it can be considered that the two groups of data are independent of each other. In other words, when filtering features, the smaller the p value, the better.
chivalue, pvalues_chi = chi2(X_fsvar,y) # The first is the chi square value and the second is the P value. It returns a one-dimensional array #To eliminate all features where the p value is greater than the set value, such as 0.05 or 0.01, the k value is set as follows: k = chivalue.shape[0] - (pvalues_chi > 0.05).sum() X_fschi = SelectKBest(chi2, k=Fill in specific k).fit_transform(X_fsvar, y) # Select k features
2.2.2 F inspection
F test, also known as ANOVA, variance homogeneity test, is a filtering method used to capture the linear relationship between each feature and label. It can do either regression or classification. Therefore, the feature is included_ selection. f_ Classif (F-test classification) and feature_selection.f_region (F-test regression) two classes. Among them, F-test classification is used for data labeled as discrete variables, while F-test regression is used for data labeled as continuous variables. It should be noted that the effect of F-test will be very stable when the data obey normal distribution. Therefore, if F-test filtering is used, the data need to be converted into a normal distribution. The use method is the same as chi square test Similarly, it needs to be used in conjunction with SelectKBest to judge what kind of K to set directly through the output statistics. The value of K can also be set through the P value.
The difference between F-test and chi square test is that F-test can be used for both regression and classification, while chi square test is only used for classification.
from sklearn.feature_selection import f_classif # Import F-test classifier F, pvalues_f = f_classif(X_fsvar,y) # Returns F and p values k = F.shape[0] - (pvalues_f > 0.05).sum() # Set K according to the returned p value X_fsF = SelectKBest(f_classif, k).fit_transform(X_fsvar, y) # Select k features
2.2. 3 mutual information method
Mutual information method is a filtering method used to capture any relationship (including linear and nonlinear relationship) between each feature and label, but it can not be used for sparse matrix. Similar to F test, mutual information method can be used for regression and classification, including two classes: feature_selection.mutual_info_classif and feature_selection.mutual_info_expression As like as two peas, F is the same as the SelectKBest test. The two classes also need to be used together with the other two.
The difference between mutual information method and F-test is that F-test can only find linear relationship, while mutual information method can find arbitrary relationship; Unlike the F-test, the mutual information method returns the p value and F value, but returns "the estimation of mutual information between each feature and target". This estimator takes a value between [0,1]. 0 means that the two variables are independent, and 1 means that the two variables are completely related.
from sklearn.feature_selection import mutual_info_classif as MIC # Import mutual information classifier result = MIC(X_fsvar,y) k = result.shape[0] - sum(result <= 0) X_fsmic = SelectKBest(MIC, k).fit_transform(X_fsvar, y)
2.2. 4 Summary
Generally, the filtering method does not perform iterative calculation on the data set, and the execution time is relatively short, but the choice of k value is given to the user, which has strong subjectivity. For the four methods of variance test, chi square test, mutual information method and F test, variance filtering is generally considered first, and then mutual information method is used to capture the correlation between labels and features. However, chi square test and F test are generally not considered because of their limitations.
2.3 embedding method
Embedding method is a method that allows the algorithm to decide which features to use. Firstly, some machine learning algorithms and models are used for training to obtain the weight coefficients of each feature, and the features are selected from large to small according to the weight coefficients. These weight coefficients often represent some contribution or importance of features to the model. Based on these importance, feature selection can be realized.
Compared with the filtering method, the result of embedding method will be more accurate to the utility of the model itself, which has a better effect on improving the effectiveness of the model. Moreover, considering the contribution of features to the model, irrelevant features (features requiring correlation filtering) and non distinguishing features (features requiring variance filtering) will be deleted because of lack of contribution to the model.
The problem of embedding method is that the calculation speed will also have a great relationship with the applied algorithm. If the algorithm with large amount of calculation and slow calculation is adopted, the embedding method itself will be very time-consuming and labor-consuming. When the algorithm itself is very complex, the calculation of the filtering method is much faster than the embedding method. In large data, the filtering method will still be given priority.
sklearn provides features_ selection. Selectfrommodel module to implement the embedding method. It has two important parameters: estimator and threshold. Estimator represents the model evaluator used, as long as it is with feature_importances_ Or coef_ Attribute, or models with I1 and I2 penalty items can be used; Threshold indicates the threshold of feature importance. Features with importance lower than this threshold will be deleted. For features_ importances_ For the model (random forest and tree model), if the importance is lower than the provided threshold parameters, these features are considered unimportant and removed. The value range of feature_imports is [0,1] , if you set a small threshold, such as 0.001, you can delete features that do not contribute to label prediction at all. If set very close to 1, only one or two features may be left.
from sklearn.feature_selection import SelectFromModel from sklearn.ensemble import RandomForestClassifier as RFC RFC_ = RFC(n_estimators =10,random_state=0) # Random forest classifier is used as model evaluator X_embedded = SelectFromModel(RFC_,threshold=0.005).fit_transform(X,y) # Below 0.005 are filtered
2.4} Wrapper method
Packaging method is also a method of feature selection and algorithm training at the same time. It is very similar to embedding method. It also depends on the choice of algorithm itself, such as coef_ Attribute or feature_importances_ Property to complete the feature selection. But the difference is that it often uses an objective function as a black box to select features, rather than entering the threshold of an evaluation index or statistic. The most typical objective function is Recursive feature elimination (recursive feature optimization, abbreviated as RFE). It is a greedy optimization algorithm designed to find the feature subset with the best performance. It repeatedly creates the model, retains the best features or eliminates the worst features in each iteration. In the next iteration, it will use the features that were not selected in the previous modeling to build the next model until all features are exhausted until. Then, it ranks the features according to the order in which it retains or eliminates the features, and finally selects the best subset.
# n_features_to_select is the number of features you want to select # step indicates the number of features to be removed in each iteration # . support_: Returns the Boolean matrix of whether all features were last selected # . ranking_ Returns the ranking of the importance of all features from sklearn.ensemble import RandomForestClassifier as RFC from sklearn.feature_selection import RFE # Objective function gradient feature elimination method RFC_ = RFC(n_estimators =10,random_state=0) # Calculating the importance of features using random forests selector = RFE(RFC_, n_features_to_select=340, step=50).fit(X, y) # Select features selector.support_.sum() selector.ranking_ X_wrapper = selector.transform(X) # Returns the selected feature
2.5 summary of feature selection
Empirically, filtering is faster, but coarser. Packaging method and embedding method are more accurate and more suitable for specific algorithm adjustment, but the amount of calculation is relatively large and the running time is long. When the amount of data is large, the variance filtering and mutual information method shall be used first, and other special features shall be added
Feature selection method. When using logistic regression, the embedding method is preferred. When using support vector machine, packaging method is preferred.
3 characteristic structure
I haven't learned it yet. I'll add later
4 feature extraction
I haven't learned it yet. I'll add later