The loss function used in this paper is the loss function constructed by KL discretization, without formula derivation; The code part is a user-defined function, not sklearn.
The loss function constructed by discrete logistic regression KL is:
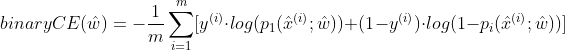
Where m is the number of samples; p_1 indicates the probability that the label is 1; y^{(i)} represents the true value of the i-th sample; x^{(i)} represents the ith sample data (including multiple features, i.e. one line of data; the last value is 1).
The derivation (gradient expression) of the loss function is:
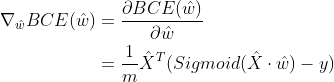
Formula derivation idea:
- BCE can be disassembled into pairs
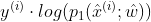 and
and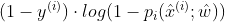 Derivative w and b respectively;
Derivative w and b respectively; - Bring the above results into
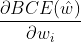 ,
,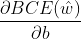
Logistic regression custom function:
def logit_gd(X,w,y):
"""
Returns the result of a logistic regression gradient descent
"""
m=X.shape[0]
return (X.T.dot(sigmoid(X.dot(w))-y))/mHow to use custom logistic regression function as prediction model? (case: label classification)
Idea:
1. Create a custom dataset
2. Use the logistic regression prediction label to draw the learning curve for model evaluation
1. Create a user-defined dataset. Use the user-defined function here to create a user-defined dataset .
def arrayGenCla(num_examples=1000,num_inputs=2,num_class=3,deg_dispersion=[4,2],bias=False):
mean_=deg_dispersion[0] # Set data mean
std_=deg_dispersion[1] # Set data variance
k=mean_*(num_class-1)/2 # Set the penalty factor to adjust the data to be distributed around the origin
cluster_l=np.empty([num_examples,1])
lf=[]
ll=[]
for i in range(num_class): # Create data for each category
data_temp=np.random.normal(i*mean_-k,std_,size=(num_examples,num_inputs))
lf.append(data_temp)
labels_temp=np.full_like(cluster_l,i)
ll.append(labels_temp)
features=np.concatenate(lf) # Merge classified data
labels=np.concatenate(ll)
if bias== True: # If the set dataset is multivariate, add "1" to the last column of features
features=np.concatenate((features,np.ones(labels.shape)),1)
return features,labels2. Use the logistic regression prediction label to draw the learning curve for model evaluation
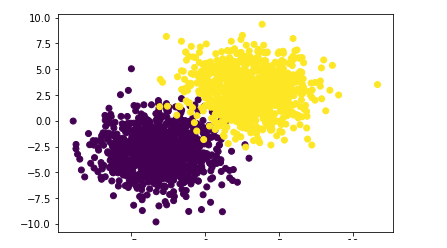
First, create a dataset using a custom function. The dataset has two labels with less overlap.
np.random.seed(9) f,l=arrayGenCla(num_class=2,deg_dispersion=[6,2],bias=True) plt.scatter(f[:,0],f[:,1],c=l) plt.show()
Then, the data set is segmented into training set and test set. The gradient descent is used to solve the parameter W, save the results after each w iteration, calculate the model performance in the training set and test set, and draw the learning curve.
Part of the code is a user-defined function, which is added behind the learning curve.
X_train,X_test,y_train,y_test=array_split(f,l,random_state=9)
X_train[:,:-1]=z_score(X_train[:,:-1])
X_test[:,:-1]=z_score(X_test[:,:-1])
np.random.seed(24)
w=np.random.randn(f.shape[1],1)
score_train=[]
score_test=[]
for i in range(num_epoch):
w=sgd_cal(X_train,w,y_train
,logit_gd
,batch_size=batch_size
,epoch=1
,lr=lr_init*lr_lambda(i))
score_train.append(logit_acc(X_train,w,y_train,thr=0.5))
score_test.append((logit_acc(X_test,w,y_test,thr=0.5)))
plt.plot(range(num_epoch),score_train,label='score_train')
plt.plot(range(num_epoch),score_test,label='score_test')
plt.legend()
plt.show()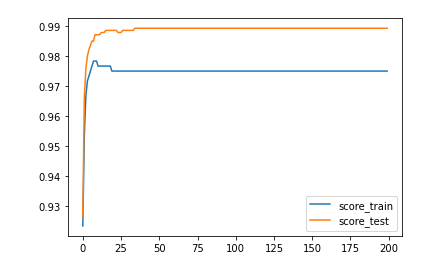
def logit_cla(yhat, thr=0.5):
"""
Return logistic regression category output function
"""
ycla = np.zeros_like(yhat)
ycla[yhat >= thr] = 1
return ycla
def logit_acc(X,w,y,thr=0.5):
"""
Return the evaluation index of logistic regression
"""
y_hat=sigmoid(X.dot(w))
y_cal=logit_cla(y_hat,thr=thr)
return(y_cal==y).mean()Conclusion: according to the learning curve, after about 25 iterations, the parameter W has been close to the optimal solution, and the model tends to be stable. The factors considered in the model setting are relatively simple, w which performs better in the test set, which is accidental.
If the data overlap more, how will this logistic regression model perform? Next, experiment again:
# Performance results of logistic regression on data sets with large degree of coincidence np.random.seed(9) f_1,l_1=arrayGenCla(num_class=2,deg_dispersion=[6,4],bias=True) plt.scatter(f_1[:,0],f_1[:,1],c=l) plt.show()
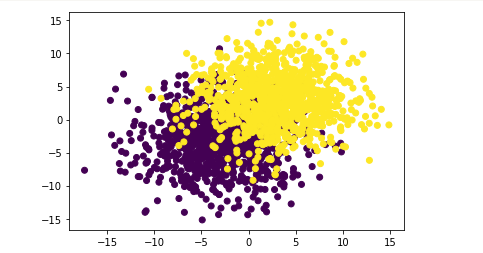
X_train,X_test,y_train,y_test=array_split(f_1,l_1,random_state=9)
X_train[:,:-1]=z_score(X_train[:,:-1])
X_test[:,:-1]=z_score(X_test[:,:-1])
np.random.seed(24)
w=np.random.randn(f.shape[1],1)
score_train=[]
score_test=[]
for i in range(num_epoch):
w=sgd_cal(X_train,w,y_train
,logit_gd
,batch_size=batch_size
,epoch=1
,lr=lr_init*lr_lambda(i)
)
score_train.append(logit_acc(X_train,w,y_train,thr=0.5))
score_test.append((logit_acc(X_test,w,y_test,thr=0.5)))
plt.plot(range(num_epoch),score_train,label='score_train')
plt.plot(range(num_epoch),score_test,label='score_test')
plt.legend()
plt.show()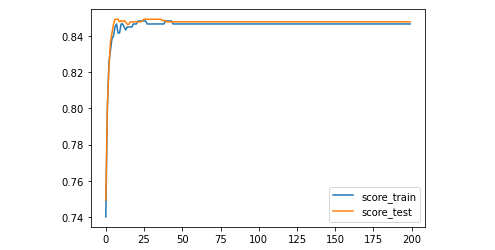
Conclusion: according to the learning curve, after 25 iterations, the model tends to be stable, and the accuracy decreases to about 0.85.