This article is edited by Markdown syntax editor.
1. DICOM module
As an open source processing software for medical images, 3d Slicer is of course inseparable from medical image data.
DICOM module is a module specially responsible for finding, acquiring, sending and receiving DICOM images.
For the description of DICOM in the official documents, see the link: https://slicer.readthedocs.io/en/latest/user_guide/modules/dicom.html
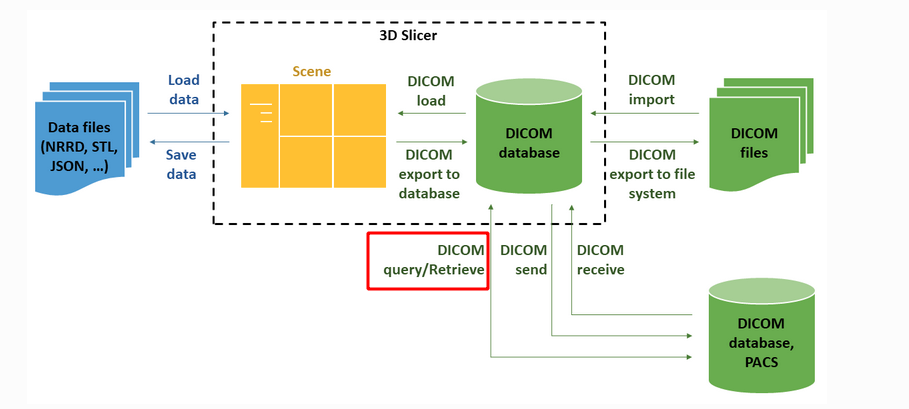
The above image introduces the process of 3d Slicer loading medical images.
3d Slicer supports loading both DICOM image data and non dcm data, such as NRRD, STL, JSON and other formats.
In addition to loading image files locally, DICOM protocol supports image transmission over the network. That is, as shown in the figure above, DICOM query/retrieve in the red box.
2. Introduction to query / retrieve function
The Query/Retrieve function corresponds to findscu and movescu/getscu in dicom protocol
| function | dcmtk instruction |
|---|---|
| Query | findscu |
| Retrieve | movescu/getscu |
In the DICOM page of 3d Slicer, there is an item: DICOM networking, which provides a function entry for DICOM image transmission through the network.
Click the "Query and retrieve" button to enter the following pop-up page.
The page shows that the default AE Title of 3d Slicer is "CTKSTORE".
The following is the information for configuring the PACS server to be connected.
Because I have built a PACS server locally with DCM4CHEE. The AE Title of PACS server is DCM4CHEE,
The Address is 192.168.109.83, the Port is 11112, and the data can be obtained through CGET.

The red box Search Options on the right is to retrieve relevant data from the PACS server through some known Patient/Study/Series information. The two columns below the search selection box look obvious. One column is the date and the other column is the modification.
However, the Name/Study/Series/ID above is not obvious. I don't know what information should be entered, and I don't know what format to enter.
Many articles about query/retrieve of 3d Slicer have been retrieved on Google, but they are all about how to query data using the following date and Modality, but there is no introduction to the above Name/Study/Series/ID.
Finally, there was no way. Only by querying the source code of 3d Slicer to find valuable information.
Because 3d Slicer is an open source project that has lasted for more than 20 years, the amount of code is quite huge. There is no better way to quickly locate relevant code in front of such a huge amount of code. I can only search by loading the code into Visual Studio Code and using the search function of the editor.
The keyword I use is "Search Options" above the search box. The first is to search in the source code, and no information was found. Later, I thought that 3d Slicer also downloaded and compiled the code of more than a dozen projects during the compilation process. Therefore, I searched in the directory of 3d Slicer bulid and found it.
The file finally located is located in the compilation directory of 3d Slicer: slicer superbuild debug:
CTK/Libs/DICOM/Core/ctkDICOMQuery.cpp function: Line 324 ~ 380 in ctkDICOMQuery:query()
/* Now, for all keys that the user provided for filtering on STUDY level,
* overwrite empty keys with value. For now, only Patient's Name, Patient ID,
* Study Description, Modalities in Study, and Study Date are used.
*/
QString seriesDescription;
foreach( QString key, d->Filters.keys() )
{
if ( key == QString("Name") && !d->Filters[key].toString().isEmpty())
{
// make the filter a wildcard in dicom style
d->Query->putAndInsertString( DCM_PatientName,
(QString("*") + d->Filters[key].toString() + QString("*")).toLatin1().data());
}
else if ( key == QString("Study") && !d->Filters[key].toString().isEmpty())
{
// make the filter a wildcard in dicom style
d->Query->putAndInsertString( DCM_StudyDescription,
(QString("*") + d->Filters[key].toString() + QString("*")).toLatin1().data());
}
else if ( key == QString("ID") && !d->Filters[key].toString().isEmpty())
{
// make the filter a wildcard in dicom style
d->Query->putAndInsertString( DCM_PatientID,
(QString("*") + d->Filters[key].toString() + QString("*")).toLatin1().data());
}
else if ( key == QString("Modalities") && !d->Filters[key].toString().isEmpty())
{
// make the filter be an "OR" of modalities using backslash (dicom-style)
QString modalitySearch("");
foreach (const QString& modality, d->Filters[key].toStringList())
{
modalitySearch += modality + QString("\\");
}
modalitySearch.chop(1); // remove final backslash
logger.debug("modalityInStudySearch " + modalitySearch);
d->Query->putAndInsertString( DCM_ModalitiesInStudy, modalitySearch.toLatin1().data() );
}
// Rememer Series Description for later series query if we go through the keys now
else if ( key == QString("Series") && !d->Filters[key].toString().isEmpty())
{
// make the filter a wildcard in dicom style
seriesDescription = "*" + d->Filters[key].toString() + "*";
}
else
{
logger.debug("Ignoring unknown search key: " + key);
}
}
if ( d->Filters.keys().contains("StartDate") && d->Filters.keys().contains("EndDate") )
{
QString dateRange = d->Filters["StartDate"].toString() +
QString("-") +
d->Filters["EndDate"].toString();
d->Query->putAndInsertString ( DCM_StudyDate, dateRange.toLatin1().data() );
logger.debug("Query on study date " + dateRange);
}
As described in the comment section of this code, the current corresponding relationship of 3d Slicer's Search Options is:

| Search Options | DICOM TAG |
|---|---|
| Name | Patient's Name |
| Study | Study Description |
| Series | Series Description |
| ID | Patient ID |
| Date | Study Date |
| Modality | Modalities in Study |
According to the above correspondence, you can enter corresponding query criteria in the corresponding Tab page of Search Options to retrieve data.
End.