Catalog
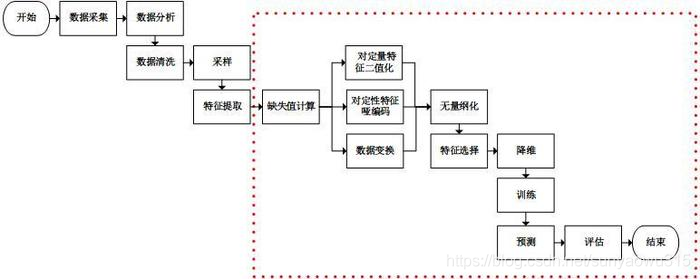
1. Overview of the process (this article is mainly the red box part)
2. Specific treatment (see red box above for specific order)
2.1 Missing value handling (delete or fill)
2.2 Data format processing (data set partition, time format, etc.)
2.3 Data sampling (oversampled or undersampled, used when uneven)
2.4 Data preprocessing (converting original data to better format standard, normalized, binary)
2.5 Feature extraction (turn text into usable_dictionary, English, DNA,TF)
2.6 Dimension reduction (filtering (low variance, correlation coefficient), PCA)
2.7 Feature Project Example: Titanic Lifetime Analysis
1. Decide what algorithm to use
2. Specific algorithm and its code
2.1 Classification Method (KNN, Logistic Regression, RF, Naive Bayesian, SVM)
2.2 Regression Method (KNN, Ridge Regression)
2.3 Clustering Method (K-means)
1. Try data and preprocessing first
2. Then select the model and adjust the model parameters
2.2 Random optimization methods (random trials)
2.3 Bayesian optimization method
2.4 Gradient-based optimization methods
2.5 Genetic algorithm (evolutionary optimization)
1. Case: Predict facebook check-in location
2. Cases: 20 categories of news
3. Case Study: Boston House Price Forecast
4. Case: Cancer Classification Prediction - Benign/Malignant Breast Cancer Prediction
7. Model preservation and loading
1. Overview of the process (this article is mainly the red box part)
2. Feature Processing Section
Introduction: The main content of this section comes from
"Feature Engineering":A Nauseous Work--Deep Understanding of Feature Engineering_wx:wu805686220-CSDN Blog
Feature Engineering Introduction_Remote-CSDN Blog
1. Overview
1.1 Some common questions
(1) There are missing values: missing values need to be supplemented (.dropna, etc.)
(2) Not in the same dimension: that is, the specifications of the features are different and cannot be compared together (standardized)
(3) Redundancy of information: some quantitative characteristics, such as interval division > If you only care about "passing" or "failing", then the score will be converted to "1" and "0" to indicate passing and failing (where > 60.)
(4) Qualitative features cannot be used directly: qualitative features need to be converted to quantitative features (such as one-hot, TF, etc.)
(5) Low information utilization: different machine learning
Algorithms and models differ in the use of information in data, so choose an appropriate model
2. Specific treatment (see red box above for specific order)
2.1 Missing value handling (delete or fill)
1) Delete missing values (dropna)
2) Filling in missing values: more commonly used, generally using the mean or majority
1 Fill with Fixed Values
A common method for missing eigenvalues is to fill them with fixed values, such as 0, -99, as in the case of -99 for this list of missing values
data['Column 1'] = data['Column 1'].fillna('-99')(2) Fill in with mean
For numeric features, the missing values can also be filled with the mean of the missing data. Here, the missing values of the feature, gray scale, are filled with the mean.
data['Column 1'] = data['Column 1'].fillna(data['Column 1'].mean()))
(3) Fill in with the majority
Similar to the mean, missing values can be populated with a majority
data['Column 1'] = data['Column 1'].fillna(data['Column 1'].mode()))
Other fills such as interpolation, KNN,RF are detailed in the original blog
2.2 Data format processing (data set partition, time format, etc.)
1) Partition of datasets
Common datasets are divided into two parts:
- Training data: for training, building models
- Test data: Used during model validation to assess whether a model is valid
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
def datasets_demo():
# Getting datasets
iris = load_iris()
print("Iris dataset:\n", iris)
print("View the dataset description:\n", iris["DESCR"])
print("View the name of the eigenvalue:\n", iris.feature_names)
print("View eigenvalues:\n", iris.data, iris.data.shape) # 150 samples
# Dataset partition X as feature Y as label
"""random_state Random number seeds, different seeds will result in different random sampling results"""
x_train, x_test, y_train, y_test = train_test_split(iris.data, iris.target, test_size=0.2, random_state=22)
return None
if __name__ == "__main__":
datasets_demo()
2) Processing of time format (mainly converting to month, day, or day of the week, and then finding what you want)
* See details https://blog.csdn.net/kobeyu652453/article/details/108894807
# Processing time data import datetime # Year, month, day years = features['year'] months = features['month'] days = features['day'] # datetime format dates = [str(int(year)) + '-' + str(int(month)) + '-' + str(int(day)) for year, month, day in zip(years, months, days)] dates = [datetime.datetime.strptime(date, '%Y-%m-%d') for date in dates] dates[:5]
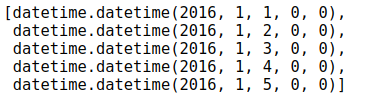
3) Tabular operations, merging or combining features
* See NP,PD tutorial for details
2.3 Data sampling (oversampled or undersampled, used when uneven)
_Oversampling of more categories/undersampling of fewer categories to balance distribution. Example: Credit card fraud, scam with too little data, use this time
def over_sample( y_origin, threshold):
y = to_one_hot(y_origin, NUM_LABELS)
y_counts = np.sum(y, axis=0)
sample_ratio = threshold / y_counts * y
sample_ratio = np.max(sample_ratio, axis=1)
sample_ratio = np.maximum(sample_ratio, 1)
index = ratio_sample(sample_ratio)
# x_token_train = [x_token_train[i] for i in index]
return y_origin[index]
def ratio_sample(ratio):
sample_times = np.floor(ratio).astype(int)
# random sample ratio < 1 (decimal part)
sample_ratio = ratio - sample_times
random = np.random.uniform(size=sample_ratio.shape)
index = np.where(sample_ratio > random)
index = index[0].tolist()
# over sample fixed integer times
row_num = sample_times.shape[0]
for row_index, times in zip(range(row_num), sample_times):
index.extend(itertools.repeat(row_index, times))
return index2.4 Data preprocessing (converting original data to better format standard, normalized, binary)
The main conversion methods are the following (the first four commonly used):
1) Standardization (very common)
Used for: data gaps between features are particularly large (e.g., deposit amounts and the number of neighborhoods around the bank, not on a scale)
from sklearn.preprocessing import StandardScaler #Standardized, returned values are standardized data StandardScaler().fit_transform(iris.data)
2) Normalization
Meaning: Sample vectors are converted into "unit vectors" when similarity is calculated by point multiplication or other kernel functions.
Under what circumstances (no) normalization is required:
- Need: Parameter-based models or distance-based models are all feature normalization.
- No: Tree-based methods do not require normalization of features, such as random forests, bagging s, boosting s, and so on.
from sklearn.preprocessing import Normalizer #Normalized, returned value is normalized data Normalizer().fit_transform(iris.data)
3) Binarization (>60 pass, marked 1)
With where or Binarizer
from sklearn.preprocessing import Binarizer #Binary, threshold set to 3, return value to binary data Binarizer(threshold=3).fit_transform(iris.data)
Other: Zoom in and out, just look at the text
2.5 Feature extraction (turn text into usable_dictionary, English, DNA,TF)
1) Dictionary feature extraction: (Dictionary to Matrix)
from sklearn.feature_extraction import DictVectorizer
def dict_demo():
data = [{'city':'Beijing', 'temperature':100},
{'city':'Shanghai', 'temperature':60},
{'city':'Shenzhen', 'temperature':30}]
# 1. Instantiate a converter class
#transfer = DictVectorizer() # Return sparse matrix
transfer = DictVectorizer(sparse=False)
# 2. Call fit_transform()
data_new = transfer.fit_transform(data)
print("data_new: \n", data_new) # Transformed
print("Feature name:\n", transfer.get_feature_names())
return None
if __name__ == "__main__":
dict_demo()Result:
data_new: [[ 0. 1. 0. 100.] [ 1. 0. 0. 60.] [ 0. 0. 1. 30.]] Feature name: ['city=Shanghai', 'city=Beijing', 'city=Shenzhen', 'temperature']
2) Text Feature Extraction (Text to Matrix)
Applying 1. Chinese and English word segmentation, then switching to matrix 2. DNA SEQS processing, bag model
2.1) English text participle
from sklearn.feature_extraction.text import CountVectorizer
def count_demo():
data = ['life is short,i like like python',
'life is too long,i dislike python']
# 1. Instantiate a converter class
transfer = CountVectorizer()
#Here's another stop_word(), which is the word to stop at
transfer1 = CountVectorizer(stop_words=['is', 'too'])
# 2. Call fit_transform
data_new = transfer.fit_transform(data)
print("data_new: \n", data_new.toarray()) # toarray to a two-dimensional array
print("Feature name:\n", transfer.get_feature_names())
return None
if __name__ == "__main__":
count_demo()
Result
data_new: [[0 1 1 2 0 1 1 0] [1 1 1 0 1 1 0 1]] Feature name: ['dislike', 'is', 'life', 'like', 'long', 'python', 'short', 'too']
2.2) Chinese word breakers
from sklearn.feature_extraction.text import CountVectorizer
import jieba
def count_chinese_demo2():
data = ['One is that today is cruel, tomorrow is cruel and the day after tomorrow is beautiful, but most of them die tomorrow evening, so everyone should not give up today.',
'We see light coming from distant galaxies millions of years ago, so when we see the universe, we are looking at its past.',
'If he knows something in only one way, he won't really know it. The secret to knowing what it really means depends on how it relates to what we know.']
data_new = []
for sent in data:
data_new.append(cut_word(sent))
print(data_new)
1,Instantiate a converter class(Call People
transfer = CountVectorizer()
2,call fit_transform(work
data_final = transfer.fit_transform(data_new)
print("data_final:\n", data_final.toarray())
print("Feature name:\n", transfer.get_feature_names())
return None
def cut_word(text):
"""
Make a Chinese participle: "I love Tian'anmen in Beijing" -> "I love Tian'anmen, Beijing"
"""
return ' '.join(jieba.cut(text))
if __name__ == "__main__":
count_chinese_demo2()
#print(cut_word('I love Tian'anmen in Beijing'))
2.3) Tf-idf Text Feature Extraction: Find Important Words and Convert to Matrix
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
import jieba
def tfidf_demo():
"""
use TF-IDF Method for text feature extraction
"""
data = ['One is that today is cruel, tomorrow is cruel and the day after tomorrow is beautiful, but most of them die tomorrow evening, so everyone should not give up today.',
'We see light coming from distant galaxies millions of years ago, so when we see the universe, we are looking at its past.',
'If he knows something in only one way, he won't really know it. The secret to knowing what it really means depends on how it relates to what we know.']
data_new = []
for sent in data:
data_new.append(cut_word(sent))
print(data_new)
1,Instantiate a converter class
transfer = TfidfVectorizer()
2,call fit_transform
data_final = transfer.fit_transform(data_new)
print("data_final:\n", data_final.toarray())
print("Feature name:\n", transfer.get_feature_names())
return None
def cut_word(text):
"""
Make a Chinese participle: "I love Tian'anmen in Beijing" -> "I love Tian'anmen, Beijing"
"""
return ' '.join(jieba.cut(text))
if __name__ == "__main__":
tfidf_demo()
#print(cut_word('I love Tian'anmen in Beijing'))
Result:
['One is that today is cruel, tomorrow is cruel and the day after tomorrow is beautiful, but most of them die tomorrow evening, so everyone should not give up today.', 'We see light coming from distant galaxies millions of years ago, so when we see the universe, we are looking at its past.', 'If he knows something in only one way, he won't really know it. The secret to knowing what it really means depends on how it relates to what we know.'] data_final: [[0.30847454 0. 0.20280347 0. 0. 0. 0.40560694 0. 0. 0. 0. 0. 0.20280347 0. 0.20280347 0. 0. 0. 0. 0.20280347 0.20280347 0. 0.40560694 0. 0.20280347 0. 0.40560694 0.20280347 0. 0. 0. 0.20280347 0.20280347 0. 0. 0.20280347 0. ] [0. 0. 0. 0.2410822 0. 0. 0. 0.2410822 0.2410822 0.2410822 0. 0. 0. 0. 0. 0. 0. 0.2410822 0.55004769 0. 0. 0. 0. 0.2410822 0. 0. 0. 0. 0.48216441 0. 0. 0. 0. 0. 0.2410822 0. 0.2410822 ] [0.12826533 0.16865349 0. 0. 0.67461397 0.33730698 0. 0. 0. 0. 0.16865349 0.16865349 0. 0.16865349 0. 0.16865349 0.16865349 0. 0.12826533 0. 0. 0.16865349 0. 0. 0. 0.16865349 0. 0. 0. 0.33730698 0.16865349 0. 0. 0.16865349 0. 0. 0. ]] Feature name: ['one kind', 'Can't,', 'No', 'before', 'understand', 'Thing', 'Today', 'Light is in', 'Millions of years', 'Issue', 'Depending on', 'only need', 'The day after tomorrow', 'Meaning', 'Gross', 'How', 'If', 'universe', 'We', 'therefore', 'give up', 'mode', 'Tomorrow', 'Galaxy', 'Night', 'Something', 'cruel', 'each', 'notice', 'real', 'Secret', 'Absolutely', 'fine', 'contact', 'Past times', 'still', 'such']
2.4) DNA SEQS treatment
1.First put seqs Data Cutting,6 A set of bases
def Kmers_funct(seq, size=6):
return [seq[x:x+size] for x in range(len(seq) - size + 1)]
2.Reuse CountVectorizer
from sklearn.feature_extraction.text import CountVectorizer
cv = CountVectorizer(ngram_range=(4,4))
X = cv.fit_transform(textsList)
3.Conversion complete, training lost2.6 Dimension reduction (filtering (low variance, correlation coefficient), PCA)
What do you mean: The process of reducing the number of random variables (characteristics) to get a set of primary variables (that is, to reduce the number of training columns)
1) Feature selection (find out important features from original features differences between human and dog: skin, height)
First: Low variance filtering (discard unimportant)
from sklearn.feature_selection import VarianceThreshold
def variance_demo():
"""
Low variance feature filtering
"""
# 1. Getting data
data = pd.read_csv('factor_returns.csv')
print('data:\n', data)
data = data.iloc[:,1:-2]
print('data:\n', data)
# 2. Instantiate a converter class
#transform = VarianceThreshold()
transform = VarianceThreshold(threshold=10)
# 3. Call fit_transform
data_new = transform.fit_transform(data)
print("data_new\n", data_new, data_new.shape)
return None
if __name__ == "__main__":
variance_demo()
Second: correlation coefficients (which are also trivial to discard, but are judged by correlation coefficients)
from sklearn.feature_selection import VarianceThreshold
from scipy.stats import pearsonr
def variance_demo():
"""
Low variance feature filter correlation coefficient
"""
# 1. Getting data
data = pd.read_csv('factor_returns.csv')
print('data:\n', data)
data = data.iloc[:,1:-2]
print('data:\n', data)
# 2. Instantiate a converter class
transform = VarianceThreshold()
transform1 = VarianceThreshold(threshold=10)
# 3. Call fit_transform
data_new = transform.fit_transform(data)
print("data_new\n", data_new, data_new.shape)
# Calculate the correlation coefficient between two variables
r = pearsonr(data["pe_ratio"],data["pb_ratio"])
print("Coefficient of correlation:\n", r)
return None
if __name__ == "__main__":
variance_demo()2)PCA
Notice mainly n_compnents: decimal indicating how much information is retained, integer how many features are reduced
from sklearn.decomposition import PCA
def pca_demo():
"""
PCA dimensionality reduction
"""
data = [[2,8,4,5], [6,3,0,8], [5,4,9,1]]
# 1. Instantiate a converter class
transform = PCA(n_components=2) # Four features down to two
# 2. Call fit_transform
data_new = transform.fit_transform(data)
print("data_new\n", data_new)
transform2 = PCA(n_components=0.95) # Keep 95% of the information
data_new2 = transform2.fit_transform(data)
print("data_new2\n", data_new2)
return None
if __name__ == "__main__":
pca_demo()
2.7 Feature Project Example: Titanic Lifetime Analysis
The above parts are commonly used by me (I compare dishes). I can see the original text in other ways.
Below is the case code
import pandas as pd
1,get data
path = "C:/DataSets/titanic.csv"
titanic = pd.read_csv(path) #1313 rows × 11 columns
# Screening Eigenvalues and Target Values
x = titanic[["pclass", "age", "sex"]]
y = titanic["survived"]
2,data processing
# 1) Treatment of missing values
x["age"].fillna(x["age"].mean(), inplace=True)
# 2) Convert to Dictionary
x = x.to_dict(orient="records")
3,Data Set Partitioning
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, random_state=22)
4,Yes x_train,test Dictionary Feature Extraction
from sklearn.feature_extraction import DictVectorizer
transfer = DictVectorizer()
x_train = transfer.fit_transform(x_train)
x_test = transfer.transform(x_test)
5.Decision Tree Estimator
from sklearn.tree import DecisionTreeClassifier, export_graphviz
estimator = DecisionTreeClassifier(criterion='entropy')
estimator.fit(x_train, y_train)
6.Model evaluation
y_predict = estimator.predict(x_test)
print("y_predict:\n", y_predict)
print("Direct required read true and predicted values:\n", y_test == y_predict) # Direct comparison
# Method 2: Calculate the accuracy
score = estimator.score(x_test, y_test) # Eigenvalues of Test Sets, Target Values of Test Sets
print("Accuracy:", score)
# Visual Decision Tree (not required)
export_graphviz(estimator, out_file='titanic_tree.dot', feature_names=transfer.get_feature_names())
import matplotlib.pyplot as plt
from sklearn.tree import plot_tree
plot_tree(decision_tree=estimator)
plt.show()
3. Common SKlearn algorithms
The main content of this article comes from 9 common algorithms for machine learning _Superior De Five-CSDN Blog _Common algorithms for machine learning
1. Decide what algorithm to use
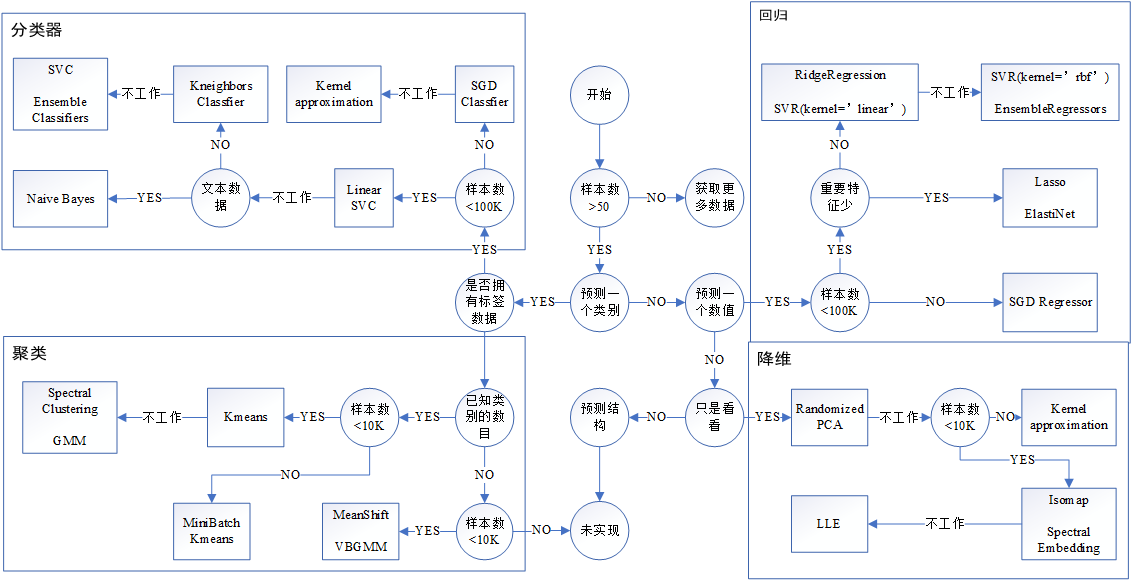
2. Specific algorithm and its code
They are basically four steps: guide (from...), modeling (knn=...), training and prediction (.fit(). predict()).
2.1 Classification Method (KNN, Logistic Regression, RF, Naive Bayesian, SVM)
2.1.1 KNN (regression, classification available)
Introduction:
The k Nearest Neighbor Classification (KNN) algorithm finds the k records closest to the new data from the training set and determines the category of the new data based on their main classification.
The algorithm involves three main points: training set, distance or similar measurement, and k size.
Advantages: Suitable for multiple classification problems
Simple, easy to understand, easy to implement, no parameter estimation, no training required
Suitable for classifying rare events (e.g., constructing loss prediction models when the loss rate is very low, e.g. less than 0.5%)
It is particularly suitable for multi-classification problems (objects with multiple class labels), such as functional classification based on genetic characteristics, where kNN performs better than SVM.
Disadvantages: High memory overhead, slower
Lazy algorithm, large amount of calculation, large memory overhead and slow score when classifying test samples
It is poorly interpretable and cannot give rules like decision trees.
Implementation code:
1.Import: Classification issues: from sklearn.neighbors import KNeighborsClassifier Regression problems: from sklearn.neighbors import KNeighborsRegressor 2.Create a model KNC = KNeighborsClassifier(n_neighbors=5) KNR = KNeighborsRegressor(n_neighbors=3) 3.train KNC.fit(X_train,y_train) KNR.fit(X_train,y_train) 4.Forecast y_pre = KNC.predict(x_test) y_pre = KNR.predict(x_test)
2.1.2 Logistic Regression (logiscic):
Introduction: Can be seen as a more accurate linear regression
The main idea of using logistic regression to classify is to establish regression formulas for classifying boundary lines based on existing data, in which the term "regression" derives from the best fit, which means to find the best set of fitting parameters.
The practice of training classifiers is to find the best fitting parameters, using the optimal algorithm. Next, the mathematical principle of this binary output classifier is introduced.
Code:
1.Import from sklearn.linear_model import LogisticRegression 2.Create a model logistic = LogisticRegression(solver='lbfgs') notes:solver Selection of parameters: "liblinear": Small-scale datasets <5~10k "lbfgs", "sag" or "newton-cg": Large-scale datasets and multi-classification problems <30k "sag": Extremely large datasets >30k 3.train logistic.fit(x_train,y_train) 4.Forecast y_pre = logistic.predict(x_train,y_train)
2.1.3 Random forests (very common)
from sklearn.datasets import load_iris
from sklearn.ensemble import RandomForestClassifier
import pandas as pd
import numpy as np
iris = load_iris()
df = pd.DataFrame(iris.data, columns=iris.feature_names)
df['is_train'] = np.random.uniform(0, 1, len(df)) <= .75
df['species'] = pd.Factor(iris.target, iris.target_names)
df.head()
train, test = df[df['is_train']==True], df[df['is_train']==False]
features = df.columns[:4]
clf = RandomForestClassifier(n_jobs=2)
y, _ = pd.factorize(train['species'])
clf.fit(train[features], y)
preds = iris.target_names[clf.predict(test[features])]
pd.crosstab(test['species'], preds, rownames=['actual'], colnames=['preds'])Short version:
# Import algorithm from sklearn.ensemble import RandomForestRegressor # modeling rf = RandomForestRegressor(n_estimators= 1000, random_state=42) # train rf.fit(train_features, train_labels) # Forecast y_pre=rf.predict(test_features)
2.1.4 Naive Bayes
Introduction: Simple Bayesian is Bayesian when variables are independent of each other. Don't be frightened by name
-
1. Gaussian (normal) distribution Naive Bayesian
-
For general classification problems
-
Use:
1.Import from sklearn.naive_bayes import GaussianNB 2.Create a model gNB = GaussianNB() 3.train gNB.fit(data,target) 4.Forecast y_pre = gNB.predict(x_test)
2. Polynomial distribution Naive Bayesian (there is also a Bernoulli distribution. Similar to this, used in small quantities)
- Suitable for text data (features represent times, such as the number of occurrences of a word)
- DNA sequences can be used as features (actually text, just AGCT)
- Commonly used for multi-classification problems
-
Use
1.Import from sklearn.naive_bayes import MultinomialNB 2.Create a model mNB = MultinomialNB() 3.Convert Character Set to Frequency from sklearn.feature_extraction.text import TfidfVectorizer #Build Tf objects first (what is Tf above) tf = TfidfVectorizer() #Train tf objects using datasets and label sets to convert tf.fit(X_train,y_train) #Text Set---->Word Frequency Set X_train_tf = tf.transform(X_train) 4.Using word frequency set to train machine learning model mNB.fit(X_train_tf,y_train) 5.Forecast #Convert Character Set to Word Frequency Set x_test = tf.transform(test_str) #Forecast mNB.predict(x_test)
2.1.5 Support Vector Machine SVM
Introduction: The idea of SVM is to find the nearest point from the hyperplane and find the optimal solution through its constraints.
Use
1.Import Handle classification issues: from sklearn.svm import SVC Handling regression issues: from sklearn.svm import SVR 2.Create a model (used in regression) SVR) svc = SVC(kernel='linear') svc = SVC(kernel='rbf') svc = SVC(kernel='poly') 3.train svc_linear.fit(X_train,y_train) svc_rbf.fit(X_train,y_train) svc_poly.fit(X_train,y_train) 4.Forecast linear_y_ = svc_linear.predict(x_test) rbf_y_ = svc_rbf.predict(x_test) poly_y_ = svc_poly.predict(x_test)
2.2 Regression Method (KNN, Ridge Regression)
2.2.1 KNN (same as above)
Ridge regression
Introduction: The improved least squares method with linear regression to avoid over-fitting and under-fitting to some extent
#1. Import from sklearn.linear_model import Ridge #2. Create a model # alpha is the reduction factor lambda, you can try the effect with your own dichotomy # If alpha is set to zero, it is a normal linear regression ridge = Ridge(alpha=0) #3. Training ridge.fit(data,target) #4. Forecast target_pre = ridge.predict(target_test)
2.2.3lasso regression
2.2.4RF (same as above)
2.2.5 Support Vector Machine SVM (same as above)
2.3 Clustering Method (K-means)
K-means (unsupervised learning)
principle
- The concept of clustering: an unsupervised learning that automatically groups similar objects into the same cluster without knowing the category beforehand.
- The K-Means algorithm is a clustering analysis algorithm. It mainly calculates the data aggregation algorithm by continuously taking the nearest mean from the seed point.
Code:
1.Import from sklearn.cluster import KMeans 2.Create a model # Building Machine Learning Objects kemans, specifying the number of classifications kmean = KMeans(n_clusters=2) 3.Training data # Note: Clustering algorithms do not have y_train kmean.fit(X_train) 4.Forecast data y_pre = kmean.predict(X_train)
IV. Assessment
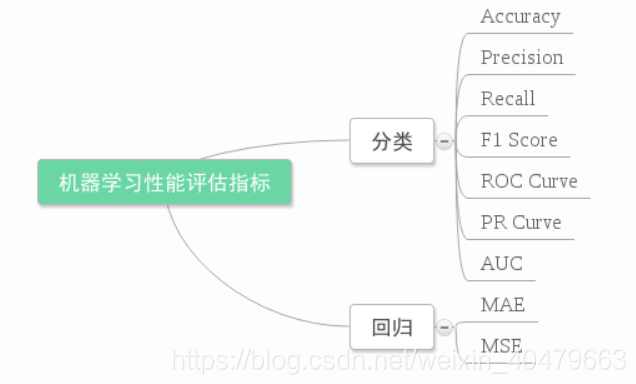
Refer to this article
5. Tuning
1. Try data and preprocessing first
Prioritize data itself and preprocessing, and work on Feature Engineering (choose more distinct features, clean data, compression, etc.)
2. Then select the model and adjust the model parameters
2.1 grid search
Introduction: Grid search is a violent method of tuning parameters by traversing all possible parameter values to get the best combination of all parameter combinations. (Just a parameter test)
Code:
#y = data['diagnosis']
#x = data.drop(['id','diagnosis','Unnamed: 32'],axis =1)
from sklearn.model_selection import train_test_split,GridSearchCV
#from sklearn.pipeline import Pipeline
#from sklearn.linear_model import LogisticRegression
#from sklearn.preprocessing import StandardScaler
#train_X,val_X,train_y,val_y = train_test_split(x,y,test_size=0.2,random_state=1)
pipe_lr = Pipeline([('scl',StandardScaler()),('clf',LogisticRegression(random_state=0))])
param_range=[0.0001,0.001,0.01,0.1,1,10,100,1000] What number to try
param_penalty=['l1','l2']
param_grid=[{'clf__C':param_range,'clf__penalty':param_penalty}]
gs = GridSearchCV(estimator=pipe_lr,
param_grid=param_grid,
scoring='f1',
cv=10,
n_jobs=-1)
gs = gs.fit(train_X,train_y)
print(gs.best_score_)
print(gs.best_params_)
2.2 Random optimization methods (random trials)
Code:
from sklearn.datasets import load_iris
from sklearn.ensemble import RandomForestRegressor
iris = load_iris()
rf = RandomForestRegressor(random_state = 42)
from sklearn.model_selection import RandomizedSearchCV
random_grid = {'n_estimators': n_estimators,
'max_features': max_features,
'max_depth': max_depth,
'min_samples_split': min_samples_split,
'min_samples_leaf': min_samples_leaf,
'bootstrap': bootstrap}
rf_random = RandomizedSearchCV(estimator = rf, param_distributions = random_grid, n_iter = 100, cv = 3, verbose=2, random_state=42, n_jobs = -1)# Fit the random search model
rf_random.fit(X,y)
#print the best score throughout the grid search
print rf_random.best_score_
#print the best parameter used for the highest score of the model.
print rf_random.best_param_The following three types do not have fixed code, depending on the specific project needs
2.3 Bayesian optimization method
2.4 Gradient-based optimization methods
2.5 Genetic algorithm (evolutionary optimization)
6. Some examples
1. Case: Predict facebook check-in location

Process analysis:
1) Get data
2) Data Processing
Purpose:
- Eigenvalue x:2<x<2.5
- Target y:1.0<y<1.5
- Time ->Years, Days, Hours and Seconds
- Filter places with fewer check-ins
3) Feature Engineering: Standardization
4) KNN algorithm prediction process
5) Model Selection and Optimization
6) Model evaluationimport pandas as pd # 1. Getting data data = pd.read_csv("./FBlocation/train.csv") #29118021 rows × 6 columns # 2. Basic Data Processing # 1) Reduce data range data = data.query("x<2.5 & x>2 & y<1.5 & y>1.0") #83197 rows × 6 columns # 2) Processing time characteristics time_value = pd.to_datetime(data["time"], unit="s") #Name: time, Length: 83197 date = pd.DatetimeIndex(time_value) data["day"] = date.day data["weekday"] = date.weekday data["hour"] = date.hour data.head() #83197 rows × 9 columns # 3) Filter places with fewer check-ins place_count = data.groupby("place_id").count()["row_id"] #2514 rows × 8 columns place_count[place_count > 3].head() data_final = data[data["place_id"].isin(place_count[place_count>3].index.values)] data_final.head() #80910 rows × 9 columns # Screening Eigenvalues and Target Values x = data_final[["x", "y", "accuracy", "day", "weekday", "hour"]] y = data_final["place_id"] # Data Set Partitioning from sklearn.model_selection import train_test_split x_train, x_test, y_train, y_test = train_test_split(x, y) from sklearn.preprocessing import StandardScaler from sklearn.neighbors import KNeighborsClassifier from sklearn.model_selection import GridSearchCV # 3. Feature Engineering: Standardization transfer = StandardScaler() x_train = transfer.fit_transform(x_train) # Standardization of training sets x_test = transfer.transform(x_test) # Test Set Standardization # 4. KNN algorithm predictor estimator = KNeighborsClassifier() # Join Grid Search and Cross Validation # Parameter preparation param_dict = {"n_neighbors": [3,5,7,9]} estimator = GridSearchCV(estimator, param_grid=param_dict, cv=5) # 10% discount, small amount of data, you can fold more estimator.fit(x_train, y_train) # 5. Model Evaluation # Method 1: Direct comparison of true and predicted values y_predict = estimator.predict(x_test) print("y_predict:\n", y_predict) print("Direct required read true and predicted values:\n", y_test == y_predict) # Direct comparison # Method 2: Calculate the accuracy score = estimator.score(x_test, y_test) # Eigenvalues of Test Sets, Target Values of Test Sets print("Accuracy:", score) # View the best parameters: best_params_ print("Optimal parameters:", estimator.best_params_) # Best result: best_score_ print("Best results:", estimator.best_score_) # Best estimator: best_estimator_ print("Best estimator:", estimator.best_estimator_) # Cross-validation results: cv_results_ print("Cross-validation results:", estimator.cv_results_)2. Cases: 20 categories of news
Step 1 Analysis
1) Get data
2) Partitioning datasets
3) Feature Engineering: Text Feature Extraction
4) Naive Bayesian predictor process
5) Model evaluation
2 Specific Code
from sklearn.model_selection import train_test_split # Partition Dataset
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import TfidfVectorizer # Text Feature Extraction
from sklearn.naive_bayes import MultinomialNB # Naive Bayes
def nb_news():
"""
Classifying News with Naive Bayesian Algorithm
:return:
"""
# 1) Get data
news = fetch_20newsgroups(subset='all')
# 2) Partitioning datasets
x_train, x_test, y_train, y_test = train_test_split(news.data, news.target)
# 3) Feature Engineering: Text Feature Extraction
transfer = TfidfVectorizer()
x_train = transfer.fit_transform(x_train)
x_test = transfer.transform(x_test)
# 4) Naive Bayesian algorithm predictor flow
estimator = MultinomialNB()
estimator.fit(x_train, y_train)
# 5) Model evaluation
y_predict = estimator.predict(x_test)
print("y_predict:\n", y_predict)
print("Direct required read true and predicted values:\n", y_test == y_predict) # Direct comparison
# Method 2: Calculate the accuracy
score = estimator.score(x_test, y_test) # Eigenvalues of Test Sets, Target Values of Test Sets
print("Accuracy:", score)
return None
if __name__ == "__main__":
nb_news()
3. Case Study: Boston House Price Forecast
1 Basic Introduction

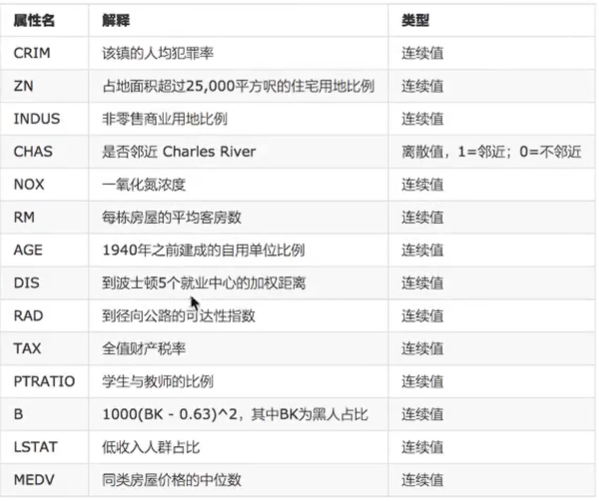
Technological process:
1) Get datasets
2) Partitioning datasets
3) Feature Engineering: No Dimension-Standardization
4) Estimator process: fit() ->model, coef_ intercept_
5) Model evaluation
2 Regression performance evaluation
Mean Square Error (MSE) Evaluation Mechanism
3 Code
from sklearn.datasets import load_boston
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LinearRegression, SGDRegressor
from sklearn.metrics import mean_squared_error
def linner1():
"""
Optimizing Methods for Normal Equations
:return:
"""
# 1) Get data
boston = load_boston()
# 2) Partitioning datasets
x_train, x_test, y_train, y_test = train_test_split(boston.data, boston.target, random_state=22)
# 3) Standardization
transfer = StandardScaler()
x_train = transfer.fit_transform(x_train)
x_test = transfer.transform(x_test)
# 4) Estimator
estimator = LinearRegression()
estimator.fit(x_train, y_train)
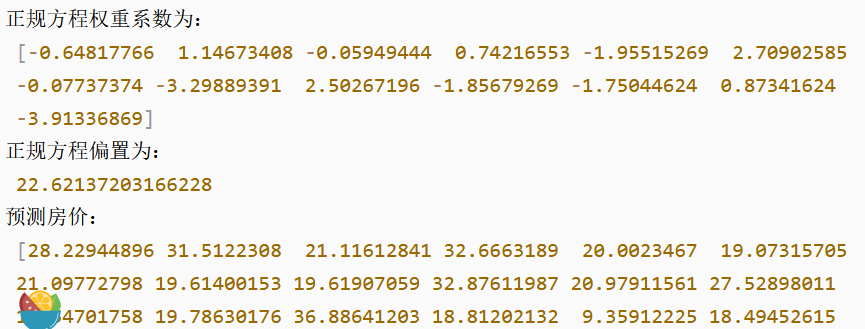
# 5) Derive the model
print("The normal equation weight factor is:\n", estimator.coef_)
print("The normal equation offset is:\n", estimator.intercept_)
# 6) Model evaluation
y_predict = estimator.predict(x_test)
print("Forecast house prices:\n", y_predict)
error = mean_squared_error(y_test, y_predict)
print("Normal Equation-The average error is:\n", error)
return None
def linner2():
"""
Optimizing method for gradient descent
:return:
"""
# 1) Get data
boston = load_boston()
print("Number of features:\n", boston.data.shape) # Several features correspond to several weight coefficients
# 2) Partitioning datasets
x_train, x_test, y_train, y_test = train_test_split(boston.data, boston.target, random_state=22)
# 3) Standardization
transfer = StandardScaler()
x_train = transfer.fit_transform(x_train)
x_test = transfer.transform(x_test)
# 4) Estimator
estimator = SGDRegressor(learning_rate="constant", eta0=0.001, max_iter=10000)
estimator.fit(x_train, y_train)
# 5) Derive the model
print("The gradient descent weight factor is:\n", estimator.coef_)
print("The gradient descent offset is:\n", estimator.intercept_)
# 6) Model evaluation
y_predict = estimator.predict(x_test)
print("Forecast house prices:\n", y_predict)
error = mean_squared_error(y_test, y_predict)
print("gradient descent-The average error is:\n", error)
return None
if __name__ == '__main__':
linner1()
linner2()
4. Case: Cancer Classification Prediction - Benign/Malignant Breast Cancer Prediction
Process analysis:
1) Get data: add names when reading
2) Data processing: processing missing values
3) Data Set Partition
4) Feature Engineering: Non-Dimensional Processing - Standardization
5) Logistic Regression Estimator
6) Model evaluation
Specific code
import pandas as pd
import numpy as np
# 1. Read Data
path = "https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data"
column_name = ['Sample code number', 'Clump Thickness', 'Uniformity of Cell Size', 'Uniformity of Cell Shape',
'Marginal Adhesion', 'Single Epithelial Cell Size', 'Bare Nuclei', 'Bland Chromatin',
'Normal Nucleoli', 'Mitoses', 'Class']
data = pd.read_csv(path, names=column_name) #699 rows × 11 columns
# 2. Treatment of missing values
# 1) Replace - "np.nan
data = data.replace(to_replace="?", value=np.nan)
# 2) Delete missing samples
data.dropna(inplace=True) #683 rows × 11 columns
# 3. Partitioning datasets
from sklearn.model_selection import train_test_split
# Screening Eigenvalues and Target Values
x = data.iloc[:, 1:-1]
y = data["Class"]
x_train, x_test, y_train, y_test = train_test_split(x, y)
# 4. Standardization
from sklearn.preprocessing import StandardScaler
transfer = StandardScaler()
x_train = transfer.fit_transform(x_train)
x_test = transfer.transform(x_test)
from sklearn.linear_model import LogisticRegression
# 5. Estimator process
estimator = LogisticRegression()
estimator.fit(x_train, y_train)
# Model parameters for logistic regression: regression coefficients and biases
estimator.coef_ # weight
estimator.intercept_ # bias
# 6. Model Evaluation
# Method 1: Direct comparison of true and predicted values
y_predict = estimator.predict(x_test)
print("y_predict:\n", y_predict)
print("Direct comparison of true and predicted values:\n", y_test == y_predict)
# Method 2: Calculate the accuracy
score = estimator.score(x_test, y_test)
print("Accuracy:\n", score)
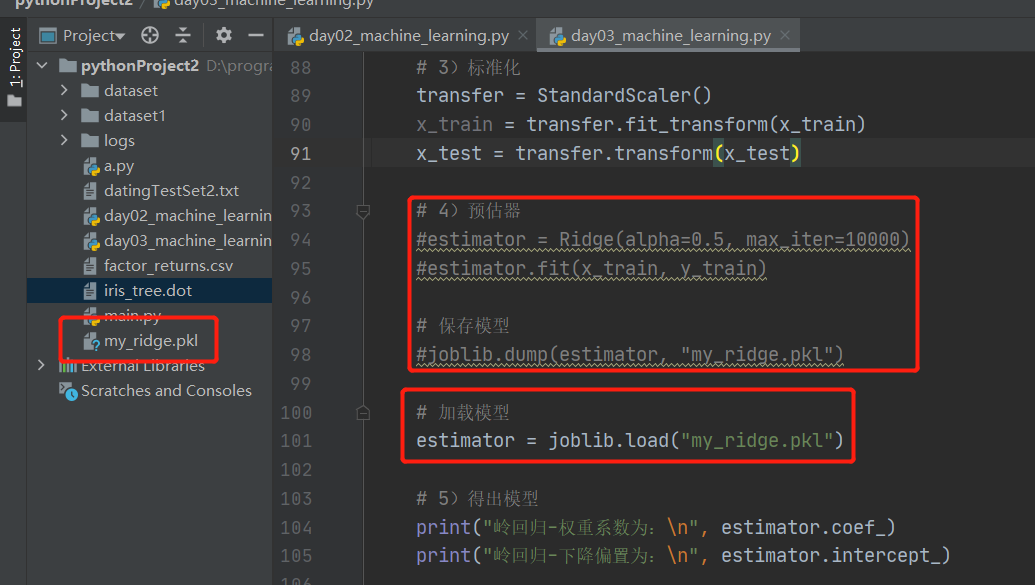
7. Model preservation and loading
import joblib
Preservation: joblib.dump(rf, 'test.pkl')
Load: estimator = joblib.load('test.pkl')Case:
1. Save the model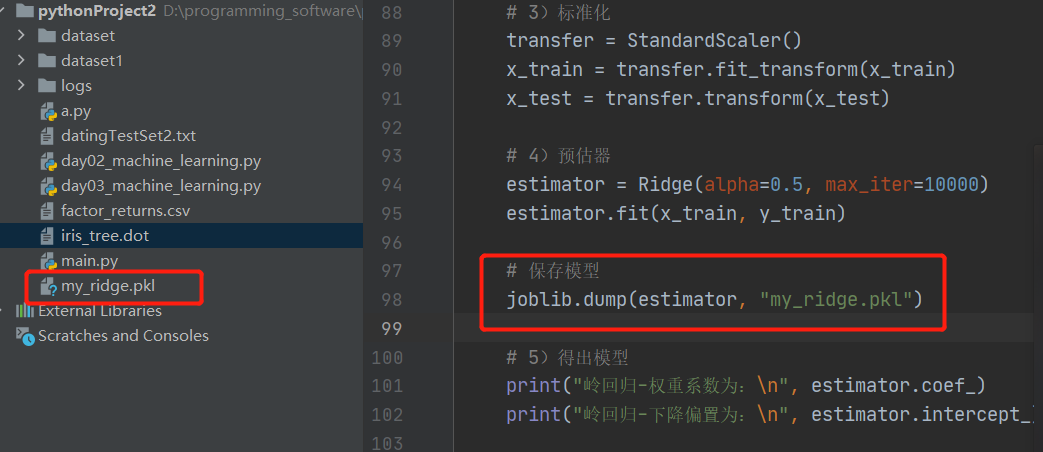
2. Loading models