Pandas Library
Introduction
Numpy performs well in vectorized numerical calculations
But when dealing with more flexible and complex data tasks:
Such as adding labels to data, dealing with missing values, grouping and PivotTables
Numpy seems powerless
The Pandas Library Based on Numpy provides advanced data structure and operation tools that make data analysis faster and simpler
11.1 object creation
11.1.1 Pandas Series object
Series is a one-dimensional array of labeled data
Creation of Series objects
General structure: PD Series(data, index=index, dtype=dtype)
Data: data, which can be a list, dictionary or Numpy array
Index: index, an optional parameter
dtype: data type; optional parameter
1 create with list
- index defaults to integer sequence
import pandas as pd data = pd.Series([1.5, 3, 4.5, 6]) print(data)
0 1.5
1 3.0
2 4.5
3 6.0
dtype: float64
- Add index
data = pd.Series([1.5, 3, 4.5, 6], index=["a", "b", "c", "d"]) print(data)
a 1.5
b 3.0
c 4.5
d 6.0
dtype: float64
- Add data type
By default, it is automatically judged from the incoming data
data = pd.Series([1, 2, 3, 4], index=["a", "b", "c", "d"], dtype=float) print(data)
a 1.0
b 2.0
c 3.0
d 4.0
dtype: float64
Note: data supports multiple data types
data = pd.Series([1, 2, "3", 4], index=["a", "b", "c", "d"]) print(data)
a 1
b 2
c 3
d 4
dtype: object
print(data["a"]) print(data["c"])
1
3
The data type can be forcibly changed
data = pd.Series([1, 2, "3", 4], index=["a", "b", "c", "d"], dtype=float) print(data)
a 1.0
b 2.0
c 3.0
d 4.0
dtype: float64
print(data["c"])
3.0
If it is not castable, an error will be reported
data = pd.Series([1, 2, "a", 4], index=["a", "b", "c", "d"], dtype=float) print(data)

2 create with one-dimensional Numpy array
import pandas as pd import numpy as np x = np.arange(5) y = pd.Series(x) print(y)
0 0
1 1
2 2
3 3
4 4
dtype: int32
3 create with dictionary
- The default key is index and the value is data
population_dict = {"BeiJing": 2154,
"ShangHai": 2424,
"ShenZhen": 1303,
"HangZhou": 981 }
population = pd.Series(population_dict)
print(population)
BeiJing 2154
ShangHai 2424
ShenZhen 1303
HangZhou 981
dtype: int64
- When creating a dictionary, if index is specified, it will be filtered from the keys in the dictionary. If it cannot be found, the value will be set to NaN
population = pd.Series(population_dict, index=["BeiJing", "HangZhou", "c", "d"]) print(population)
BeiJing 2154.0
HangZhou 981.0
c NaN
d NaN
dtype: float64
4. When data is scalar
print(pd.Series(5, index=[100, 200, 300]))
100 5
200 5
300 5
dtype: int64
11.1. 2. Pandas dataframe object
DataFrame is a multidimensional array of labeled data
Creation of DataFrame object
General structure: PD DataFrame(data, index=index, columns=columns)
Data: data, which can be a list, dictionary or Numpy array
Index: index, an optional parameter
columns: column label, optional parameter
1 create through Series objects
population_dict = {"BeiJing": 2154,
"ShangHai": 2424,
"ShenZhen": 1303,
"HangZhou": 981}
population = pd.Series(population_dict)
print(pd.DataFrame(population))

print(pd.DataFrame(population, columns=["population"]))
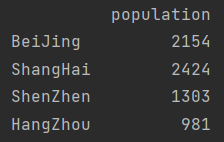
2 create through Series object dictionary
population_dict = {"BeiJing": 2154,
"ShangHai": 2424,
"ShenZhen": 1303,
"HangZhou": 981}
population = pd.Series(population_dict)
GDP_dict = {"BeiJing": 30320,
"ShangHai": 32680,
"ShenZhen": 24222,
"HangZhou": 13468}
GDP = pd.Series(GDP_dict)
print(pd.DataFrame({"population": population,
"GDP": GDP}))
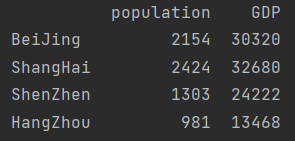
Note: if the quantity is insufficient, it will be supplemented automatically
print(pd.DataFrame({"population": population,
"GDP": GDP,
"Country": "China"}))
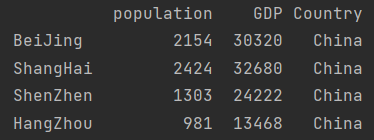
3 create through dictionary list object
- The dictionary index is used as index and the dictionary key is used as columns
data = [{"a": i, "b": 2*i} for i in range(3)]
print(data)
print(pd.DataFrame(data))
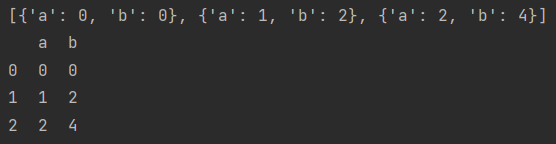
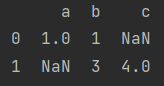
- Keys that do not exist will default to NaN
data = [{"a": 1, "b": 1}, {"b":3, "c": 4}]
print(pd.DataFrame(data))
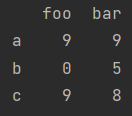
4 created by Numpy two-dimensional array
print(pd.DataFrame(np.random.randint(10, size=(3, 2)), columns=["foo", "bar"], index=["a", "b", "c"]))
11.2 properties of dataframe
11.2. 1 Properties
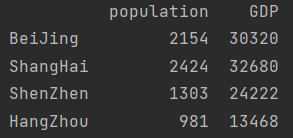
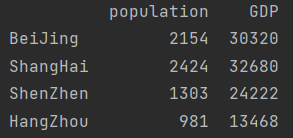
data = pd.DataFrame({"population":population, "GDP": GDP})
print(data)
1. df.values returns the data represented by the numpy array
print(data.values)
[[ 2154 30320]
[ 2424 32680]
[ 1303 24222]
[ 981 13468]]
2. df.index returns the row index
print(data.index)
Index(['BeiJing', 'ShangHai', 'ShenZhen', 'HangZhou'], dtype='object')
3. df.columns returns the column index
print(data.columns)
Index(['population', 'GDP'], dtype='object')
4. df.shape shape
print(data.shape)
(4, 2)
5. pd.size size
print(data.size)
8
6. pd.dtypes returns the data type of each column
print(data.dtypes)
population int64
GDP int64
dtype: object
11.2. 2 index
1. Get columns
- Dictionary type
print(data["population"])
BeiJing 2154
ShangHai 2424
ShenZhen 1303
HangZhou 981
Name: population, dtype: int64
print(data[["GDP", "population"]]) # Multiple columns are obtained in list form
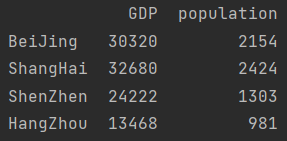
- Object attribute
print(data.GDP)
BeiJing 30320
ShangHai 32680
ShenZhen 24222
HangZhou 13468
Name: GDP, dtype: int64
2. Get row
- Absolute index DF loc
print(data.loc["BeiJing"])
population 2154
GDP 30320
Name: BeiJing, dtype: int64
print(data.loc[["BeiJing", "HangZhou"]])
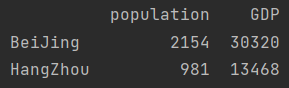
- Relative index
print(data.iloc[0])
population 2154
GDP 30320
Name: BeiJing, dtype: int64
print(data.iloc[[1, 3]])
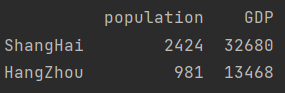
3. Get scalar
print(data.loc["BeiJing", "GDP"])
30320
print(data.iloc[0, 1])
30320
print(data.values[0][1])
30320
4. Index of series objects
print(type(data.GDP))
<class 'pandas.core.series.Series'>
print(GDP["BeiJing"])
30320
11.2. 3 slice
dates = pd.date_range(start='2019-01-01', periods=6) print(dates)
DatetimeIndex(['2019-01-01', '2019-01-02', '2019-01-03', '2019-01-04',
'2019-01-05', '2019-01-06'],
dtype='datetime64[ns]', freq='D')
df = pd.DataFrame(np.random.randn(6, 4), index=dates, columns=["A", "B", "C", "D"]) print(df)
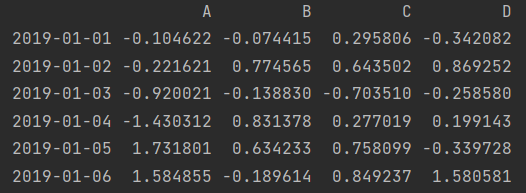
1. Row slicing
print(df["2019-01-01": "2019-01-03"]) print(df.loc["2019-01-01": "2019-01-03"]) print(df.iloc[0:3]) # End position not included
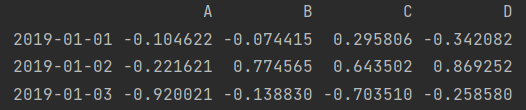
2. Column slicing
print(df.loc[:, "A": "C"]) print(df.iloc[:, 0: 3])
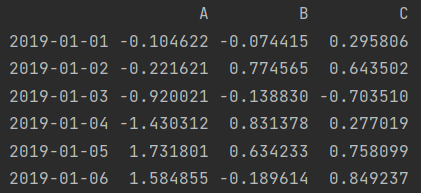
3. Various values
- Row and column slicing at the same time
print(df.loc["2019-01-02": "2019-01-03", "C": "D"]) # Row and column simultaneous slicing print(df.iloc[1: 3, 2:])
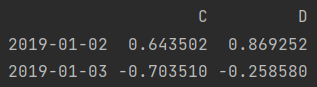
- Row slice, column scatter value
print(df.loc["2019-01-04": "2019-01-06", ["A", "C"]]) # Row slice, column scatter value print(df.iloc[3:, [0, 2]])
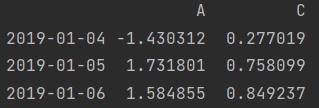
- Row scatter value, column slice
print(df.loc[["2019-01-04", "2019-01-06"], "C": "D"]) # Row scatter value, column slice print(df.iloc[[3, 5], 2:])
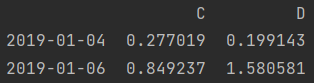
- Row and column values are scattered
print(df.loc[["2019-01-04", "2019-01-06"], ["C", "D"]]) # Row and column values are scattered print(df.iloc[[3, 5], [2, 3]])

11.2. 4 Boolean index
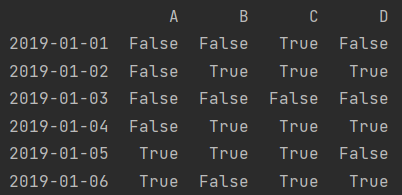
print(df)

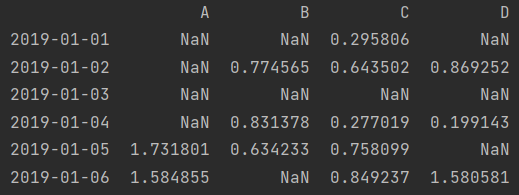
print(df > 0)
print(df[df > 0])
print(df.A > 0)
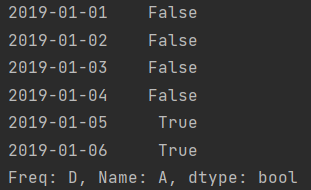
print(df[df.A > 0])
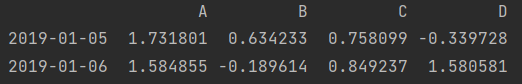
- isin() method
df2 = df.copy() df2['E'] = ['one', 'one', 'two', 'three', 'four', 'three'] print(df2)
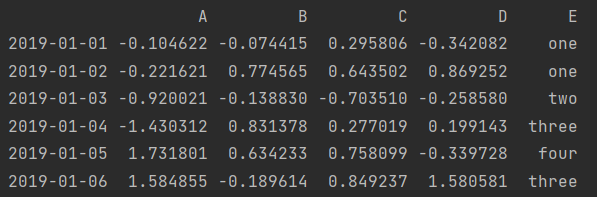
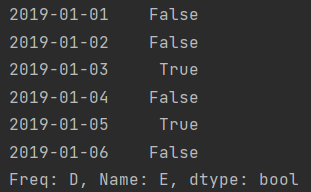
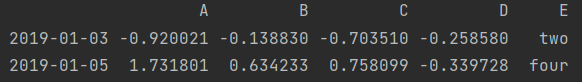
ind = df2['E'].isin(["two", "four"]) print(ind) print(df2[ind])
11.2. 5 assignment
print(df)

- Add new column to DataFrame
s1 = pd.Series([1, 2, 3, 4, 5, 6], index=pd.date_range('20190101', periods=6))
print(s1)
2019-01-01 1
2019-01-02 2
2019-01-03 3
2019-01-04 4
2019-01-05 5
2019-01-06 6
Freq: D, dtype: int64
df['E'] = s1 print(df)
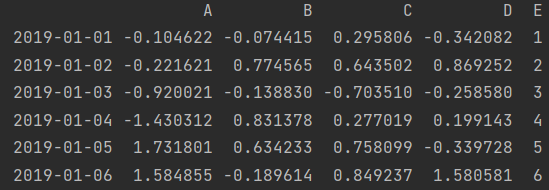
- Modify assignment
df.loc["2019-01-01", "A"] = 0 print(df)
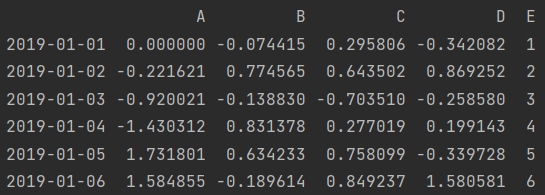
df.iloc[0, 1] = 0 print(df)
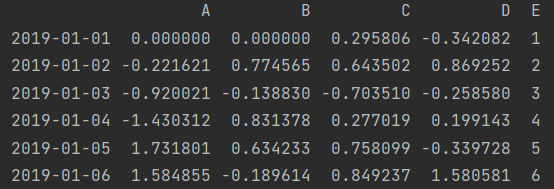
df["D"] = np.array([5]*len(df)) # It can be simplified to df["D"] = 5 print(df)
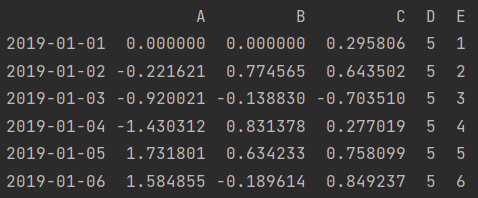
- Modify index and columns
df.index = [i for i in range(len(df))] df.columns = [i for i in range(df.shape[1])] print(df)

11.3 numerical calculation and statistical analysis
11.3. 1. View data
dates = pd.date_range(start='2019-01-01', periods=6) df = pd.DataFrame(np.random.randn(6, 4), index=dates, columns=["A", "B", "C", "D"]) print(df)
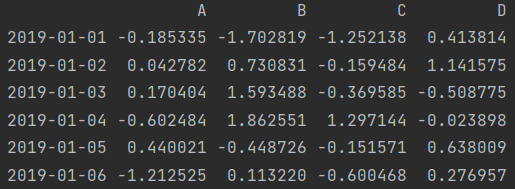
1. View the previous line
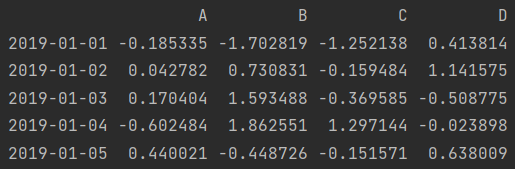
print(df.head()) # Default 5 lines
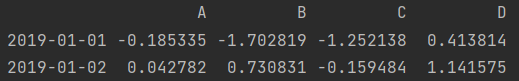
print(df.head(2))
2. View the following lines
print(df.tail()) # Default 5 lines
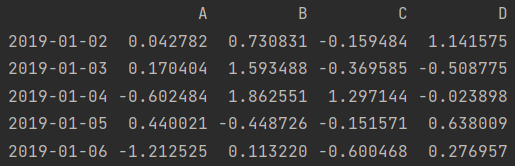
print(df.tail(3))
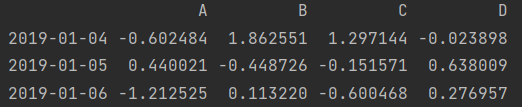
3. View general information
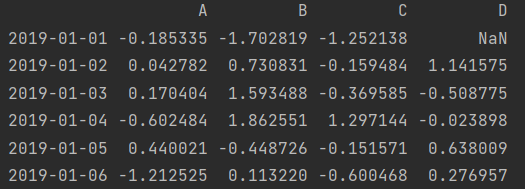
In order to view the overall information, we first change one of the elements to NaN
df.iloc[0, 3] = np.nan print(df)
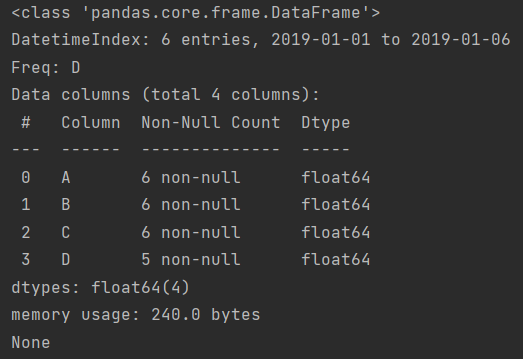
print(df.info())
11.3.2 Numpy general function is also applicable to Pandas
1 vectorization operation
x = pd.DataFrame(np.arange(4).reshape(1, 4)) print(x)
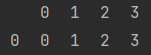
print(x + 5)

print(np.exp(x))
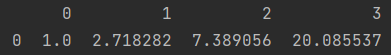
y = pd.DataFrame(np.arange(4, 8).reshape(1, 4)) print(x * y)

2 matrix operation
np.random.seed(42) x = pd.DataFrame(np.random.randint(10, size=(10, 10))) print(x)
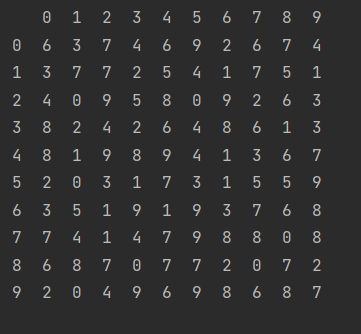
- Transpose
z = x.T print(z)
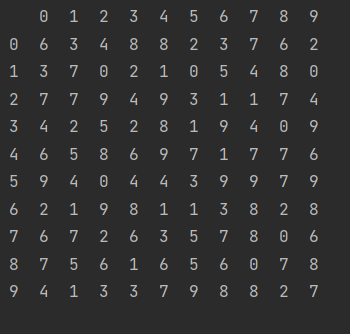
- Matrix multiplication
np.random.seed(1) y = pd.DataFrame(np.random.randint(10, size=(10, 10))) print(x.dot(y))
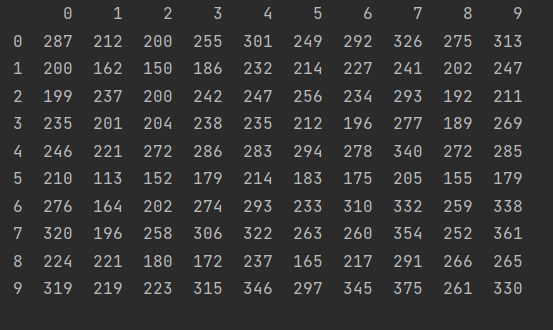
We can use the% timeit method to compare the time required for the next two kinds of multiplication
%timeit x.dot(y) %timeit np.dot(x, y)
NP can be found Dot (x, y) takes less time
- Perform the same operation, comparison between Numpy and Pandas
In general, pure computing is faster in Numpy
Numpy focuses more on calculation and Pandas focuses more on data analysis
3 broadcast operation
np.random.seed(42)
x = pd.DataFrame(np.random.randint(10, size=(3, 3)), columns=list("ABC"))
print(x)

- Spread by line
print(x.iloc[0])
A 6
B 3
C 7
Name: 0, dtype: int32
print(x/x.iloc[0])
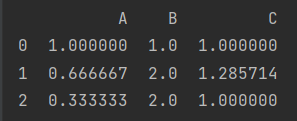
- Column propagation
print(x.div(x.A, axis=0)) # add sub div mul
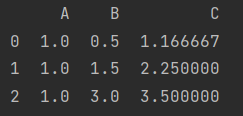
Here, if axis is set to 1, it means propagation by column. You can also explicitly write out that axis=0 means propagation by row.
11.3. 3 new usage
1 Index alignment
np.random.seed(42)
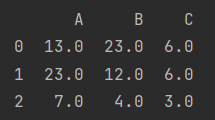
A = pd.DataFrame(np.random.randint(0, 20, size=(2, 2)), columns=list("AB"))
B = pd.DataFrame(np.random.randint(0, 10, size=(3, 3)), columns=list("ABC"))
print(A)
print(B)
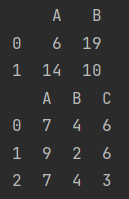
- Pandas will automatically align the indexes of the two objects, and NP. Is used for values without Nan representation
print(A+B)
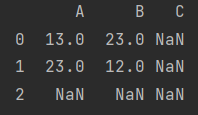
- The default value can be filled with fill value
print(A.add(B, fill_value=0))
2 statistical correlation
- Data type statistics
y = np.random.randint(3, size=20) print(y)
[2 0 2 2 0 0 2 1 2 2 2 2 0 2 1 0 1 1 1 1]
print(np.unique(y))
[0 1 2]
from collections import Counter print(Counter(y))
Counter({2: 9, 1: 6, 0: 5})
y1 = pd.DataFrame(y, columns=["A"]) print(y1)
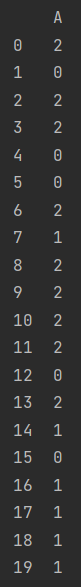
print(np.unique(y1))
[0 1 2]
print(y1["A"].value_counts())
2 9
1 6
0 5
Name: A, dtype: int64
- Generate new results and sort them
population_dict = {"BeiJing": 2154,
"ShangHai": 2424,
"ShenZhen": 1303,
"HangZhou": 981}
population = pd.Series(population_dict)
GDP_dict = {"BeiJing": 30320,
"ShangHai": 32680,
"ShenZhen": 24222,
"HangZhou": 13468}
GDP = pd.Series(GDP_dict)
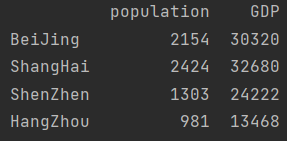
city_info = pd.DataFrame({"population": population, "GDP": GDP})
print(city_info)
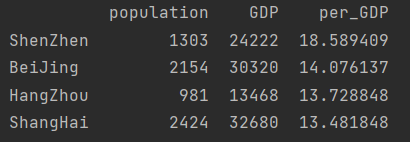
city_info["per_GDP"] = city_info["GDP"]/city_info["population"] print(city_info)

sort ascending
print(city_info.sort_values(by="per_GDP"))

sort descending
print(city_info.sort_values(by="per_GDP", ascending=False))
Sort by axis
data = pd.DataFrame(np.random.randint(20, size=(3, 4)), index=[2, 1, 0], columns=list("CBAD"))
print(data)
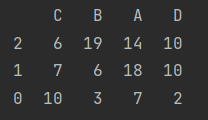
Row sorting
print(data.sort_index())
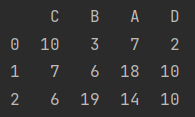
Column sorting
print(data.sort_index(axis=1))
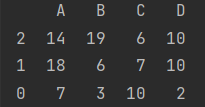
- statistical method
df = pd.DataFrame(np.random.normal(2, 4, size=(6, 4)), columns=list("ABCD"))
print(df)
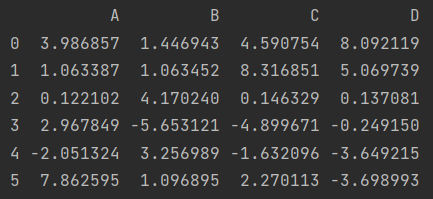
Non empty number
df.count()
A 6
B 6
C 6
D 6
dtype: int64
Summation (default summation of columns)
df.sum()
A 13.951465
B 5.381398
C 8.792280
D 5.701582
dtype: float64
df.sum(axis=1) # Sum of rows
0 18.116673
1 15.513429
2 4.575753
3 -7.834093
4 -4.075646
5 7.530610
dtype: float64
Max min
df.min() # Default minimum value of each column
A -2.051324
B -5.653121
C -4.899671
D -3.698993
dtype: float64
df.max(axis=1) # Maximum value of each row after shaft replacement
0 8.092119
1 8.316851
2 4.170240
3 2.967849
4 3.256989
5 7.862595
dtype: float64
print(df.idxmax()) # Maximum coordinates
A 5
B 2
C 1
D 0
dtype: int64
mean value
df.mean()
A 2.325244
B 0.896900
C 1.465380
D 0.950264
dtype: float64
variance
df.var()
A 11.887325
B 11.911567
C 21.841270
D 22.569365
dtype: float64
standard deviation
df.std()
A 3.447800
B 3.451314
C 4.673464
D 4.750723
dtype: float64
median
df.median()
A 2.015618
B 1.271919
C 1.208221
D -0.056035
dtype: float64
Mode
data = pd.DataFrame(np.random.randint(5, size=(10, 2)), columns=list("AB"))
print(data)
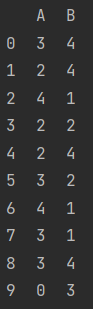
data.mode()

75% quantile
df.quantile(0.75)
A 3.732105
B 2.804478
C 4.010594
D 3.836574
Name: 0.75, dtype: float64
catch all in one draft
df.describe()
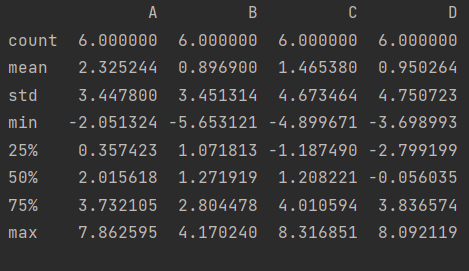
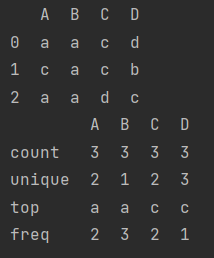
The character type also has a describe
data_2 = pd.DataFrame([["a", "a", "c", "d"],
["c", "a", "c", "b"],
["a", "a", "d", "c"]], columns=list("ABCD"))
print(data_2)
print(data_2.describe())
Correlation coefficient and covariance
print(df.corr())
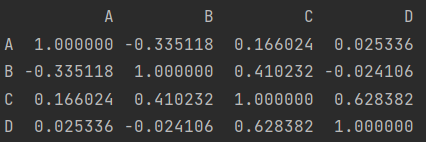
print(df.corrwith(df["A"]))
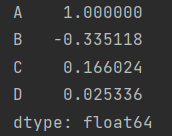
Custom output
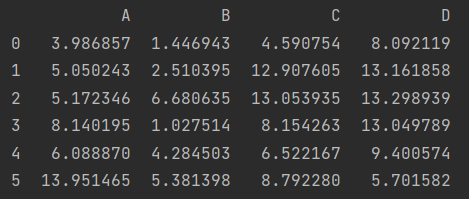
Usage of apply(method): use the method to perform corresponding operations on each column by default
df.apply(np.cumsum)
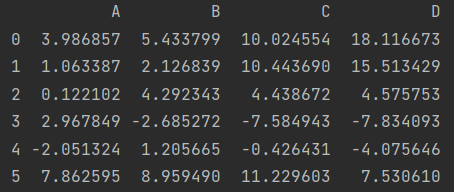
print(df.apply(np.cumsum, axis=1))
df.apply(sum)
A 13.951465
B 5.381398
C 8.792280
D 5.701582
dtype: float64
df.apply(lambda x: x.max()-x.min())
A 9.913920
B 9.823361
C 13.216523
D 11.791112
dtype: float64
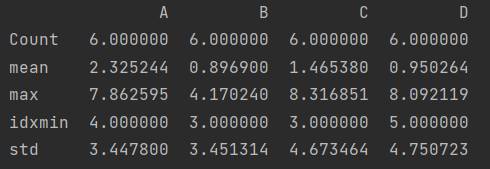
def my_describe(x):
return pd.Series([x.count(), x.mean(), x.max(), x.idxmin(), x.std()], \
index=["Count", "mean", "max", "idxmin", "std"])
print(df.apply(my_describe))
11.4 missing value handling
11.4. 1 missing value found
import pandas as pd
import numpy as np
data = pd.DataFrame(np.array([[1, np.nan, 2],
[np.nan, 3, 4],
[5, 6, None]]), columns=["A", "B", "C"])
print(data)
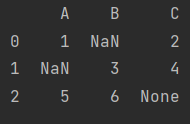
Note: there are None, string, etc. the data type is changed to object, which consumes more resources than int and float
data.dtypes
A object
B object
C object
dtype: object
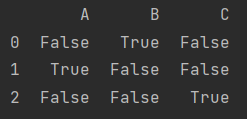
data.isnull()
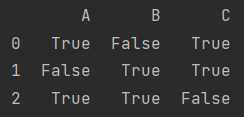
data.notnull()
11.4. 2 delete missing values
data = pd.DataFrame(np.array([[1, np.nan, 2, 3],
[np.nan, 4, 5, 6],
[7, 8, np.nan, 9],
[10, 11, 12, 13]]), columns=["A", "B", "C", "D"])
print(data)
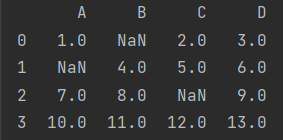
Note: NP Nan is a special floating point number
data.dtypes
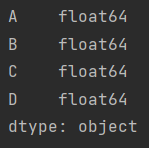
1. Delete the whole line
data.dropna()
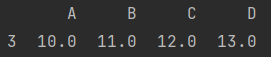
2. Delete the whole column
data.dropna(axis="columns")
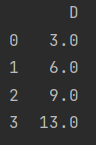
data["D"] = np.nan print(data)
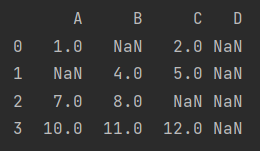
data.dropna(axis="columns", how="all") # Delete when all elements are missing. any means delete when there is a missing element
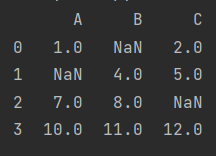
11.4. 3 fill in missing values
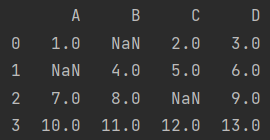
data.fillna(value=5)
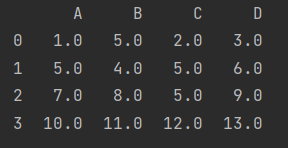
- Replace with mean
fill = data.mean() print(data.fillna(value=fill))
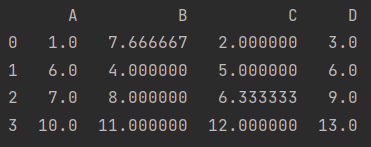
If all average values are used instead, the average value can be taken after stack leveling
fill = data.stack().mean() print(data.fillna(value=fill))
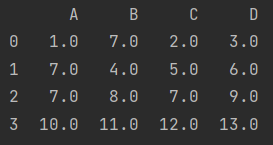
11.5 consolidated data
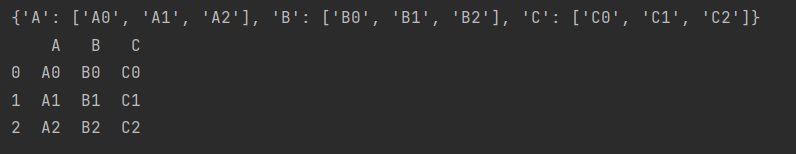
- Construct a function to produce DataFrame
def make_df(cols, ind):
"""A simple DataFrame"""
data = {c: [str(c)+str(i) for i in ind] for c in cols}
print(data)
return pd.DataFrame(data, ind)
make_df("ABC", range(3))
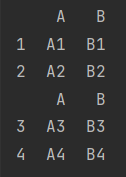
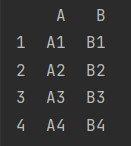
- Vertical merge
If the row labels overlap, the overlapping row labels are retained
df_1 = make_df("AB", [1, 2])
df_2 = make_df("AB", [3, 4])
print(df_1)
print(df_2)
print(pd.concat([df_1, df_2]))
- Horizontal merge
If the column labels overlap, the overlapping column labels are retained
df_3 = make_df("AB", [0, 1])
df_4 = make_df("CD", [0, 1])
print(df_3)
print(df_4)
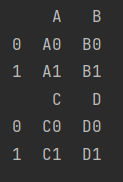
print(pd.concat([df_3, df_4], axis=1))
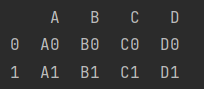
- Index overlap
When labels overlap, we sometimes want labels to be rearranged, so we need to add the parameter ignore_index=True
df_5 = make_df("AB", [1, 2])
df_6 = make_df("AB", [1, 2])
print(df_5)
print(df_6)
print(pd.concat([df_5, df_6], ignore_index=True))
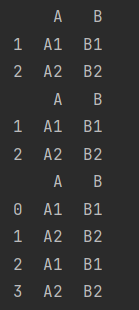
The same is true for column overlap. Add the parameter axis=1
- Align merge()
merge() can be used to merge when there are corresponding same values (common columns)
df_9 = make_df("AB", [1, 2])
df_10 = make_df("BC", [1, 2])
print(df_9)
print(df_10)
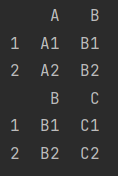
We can see column B in df_9 and DF_ The correspondence in 10 is equal. We want column B to remain unchanged when merging
print(pd.merge(df_9, df_10))
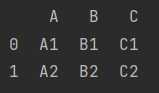
[example] merge city information
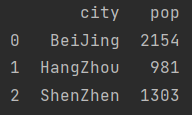
population_dict = {"city": ("BeiJing", "HangZhou", "ShenZhen"),
"pop": (2154, 981, 1303)}
population = pd.DataFrame(population_dict)
print(population)
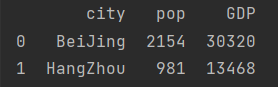
GDP_dict = {"city": ("BeiJing", "ShangHai", "HangZhou"),
"GDP": (30320, 32680, 13468)}
GDP = pd.DataFrame(GDP_dict)
print(GDP)

city_info = pd.merge(population, GDP) print(city_info)
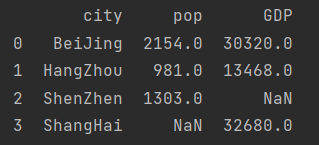
If you don't want to abandon Shanghai and Shenzhen, you need to add parameters
city_info = pd.merge(population, GDP, how="outer") print(city_info)
11.6 grouping and PivotTables
df = pd.DataFrame({"key": ["A", "B", "C", "C", "B", "A"],
"data1": range(6),
"data2": np.random.randint(0, 10, size=6)})
print(df)
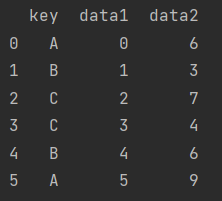
11.6. 1 grouping
- Delay calculation (save first and wait for processing)
df.groupby("key")
<pandas.core.groupby.generic.DataFrameGroupBy object at 0x000001EDFF4444C0>
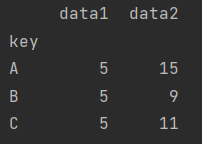
df.groupby("key").sum()
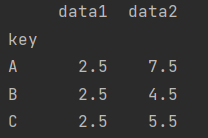
df.groupby("key").mean()
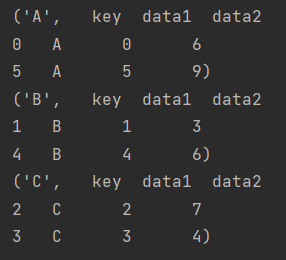
So what exactly is groupby?
for i in df.groupby("key"):
print(str(i))
- Value by column
df.groupby("key")["data2"].sum()
key
A 15
B 9
C 11
Name: data2, dtype: int32
- Iterate by group
for data, group in df.groupby("key"):
print("{0:5} shape={1}".format(data, group.shape))
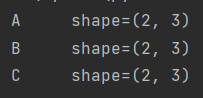
- Call method
df.groupby("key")["data1"].describe()
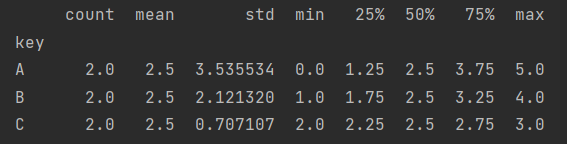
- Support for more complex operations
df.groupby("key").aggregate(["min", "median", "max"])
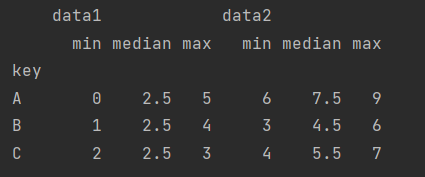
- filter
np.random.seed(1)
df = pd.DataFrame({"key": ["A", "B", "C", "C", "B", "A"],
"data1": range(6),
"data2": np.random.randint(0, 10, size=6)})
print(df)
def filter_func(x):
return x["data2"].std() > 3
print(df.groupby("key")["data2"].std())
print(df.groupby("key").filter(filter_func))
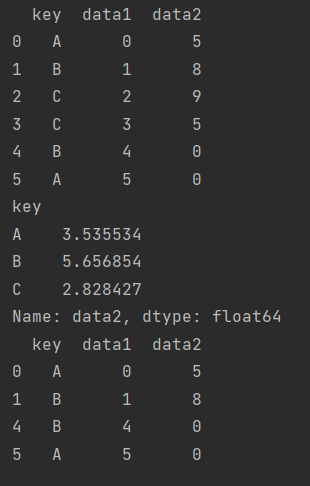
- transformation
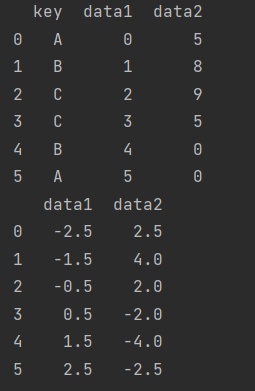
df = pd.DataFrame({"key": ["A", "B", "C", "C", "B", "A"],
"data1": range(6),
"data2": np.random.randint(0, 10, size=6)})
print(df)
print(df.groupby("key").transform(lambda x: x - x.mean()))
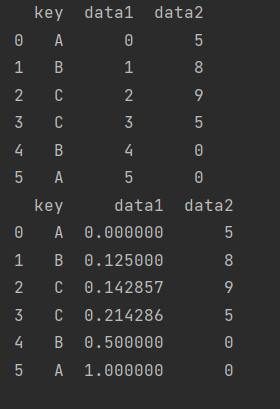
- apply method
def norm_by_data2(x):
x["data1"] /= x["data2"].sum()
return x
print(df.groupby("key").apply(norm_by_data2))
- Set list and array as grouping key
print(df) L = [0, 1, 0, 1, 2, 0] print(df.groupby(L).sum())

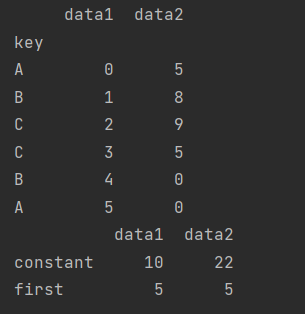
- Mapping indexes to groups with dictionaries
df2 = df.set_index("key")
print(df2)
mapping = {"A": "first", "B": "constant", "C": "constant"}
print(df2.groupby(mapping).sum())
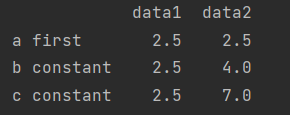
- Any Python function
print(df2.groupby(str.lower).mean())

- A list of valid values
print(df2.groupby([str.lower, mapping]).mean())
[example 1] planetary observation data processing
import seaborn as sns
planets = sns.load_dataset('planets')
print(planets.shape) # Get shape
print(planets.head()) # View the first five lines
print(planets.describe()) # statistical information
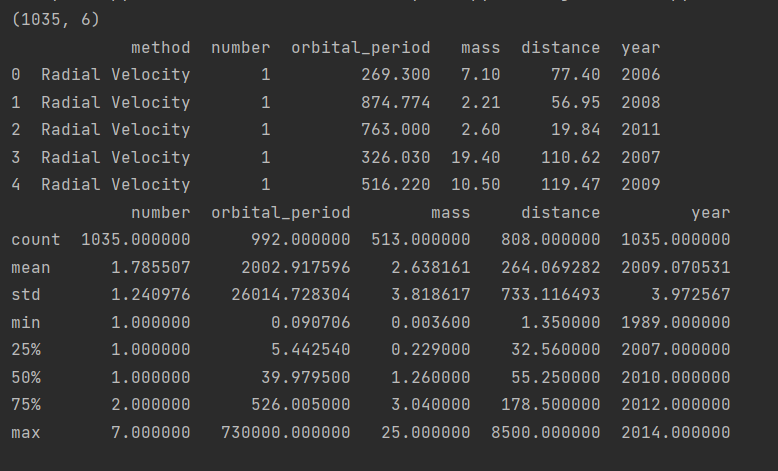
Now we hope to realize the observation quantity in different times (decade, unit) under different observation methods by grouping.
First, we should define the era.
decade = 10 * (planets["year"]//10) print(decade.head()) decade = decade.astype(str) + "s" decade.name = "decade" print(decade.head())
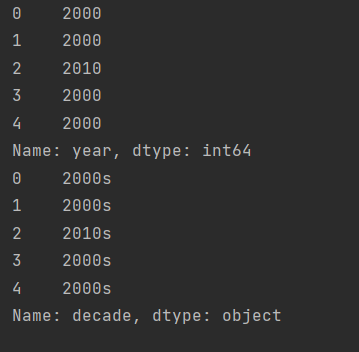
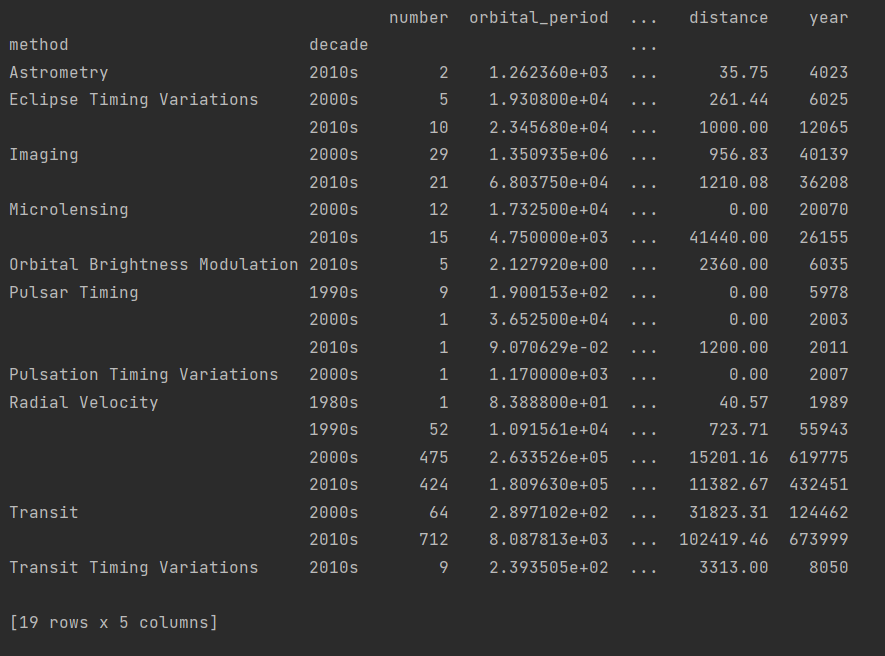
After definition, we group the method and decade.
print(planets.groupby(["method", decade]).sum())
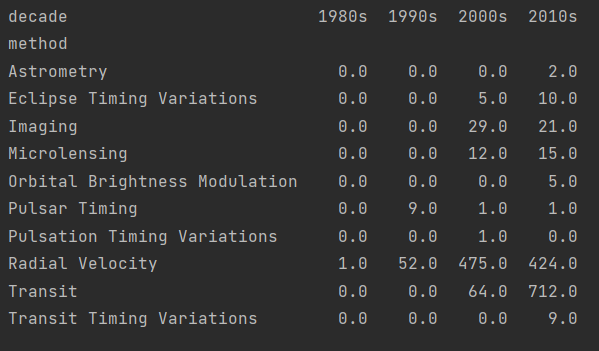
In fact, we only need the data in the column number, so we can propose number separately, expand it with unstack to make it more beautiful, and finally fill the missing value with 0 with fillna.
print(planets.groupby(["method", decade])["number"].sum().unstack().fillna(0))
11.6. 2 PivotTable report
[example 2] analysis of Titanic passenger data
import seaborn as sns
titanic = sns.load_dataset("titanic")
print(titanic.head())
print(titanic.describe())

Now we want to get the survival rate of different genders.
print(titanic.groupby("sex")["survived"].mean())
sex
female 0.742038
male 0.188908
Name: survived, dtype: float64
The index "served" is expressed as Series in one bracket and DataFrame in two brackets
print(titanic.groupby("sex")[["survived"]].mean())
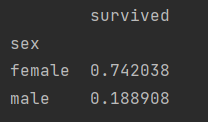
More specifically, we want to see the survival of different cabins under different genders.
print(titanic.groupby(["sex", "class"])["survived"].aggregate("mean").unstack())
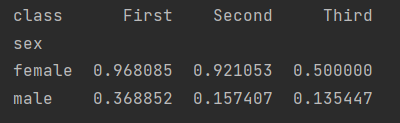
We found that writing by groupby alone is complex and unreadable, but Pandas provides a PivotTable method. Let's take a look at how to use PivotTable to achieve the same function.
print(titanic.pivot_table("survived", index="sex", columns="class"))
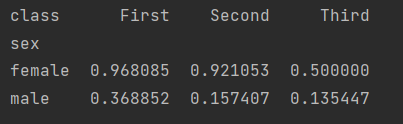
We can find that we can use pivot directly_ The table method returns the average value of data by default.
We can also add other parameters to get different results.
aggfunc to change the method and margins to set the overview.
print(titanic.pivot_table("survived", index="sex", columns="class", aggfunc="mean", margins=True))
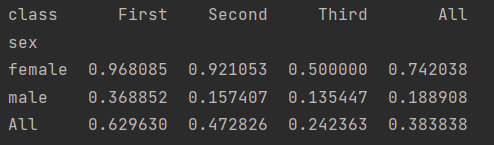
When we use pivot_ If you want to process multiple types of data, you can select the corresponding methods in aggfuc. For example, in this example, we not only want to get the number of survival, but also need the average ticket price.
print(titanic.pivot_table(index="sex", columns="class", aggfunc={"survived": "sum", "fare": "mean"}))
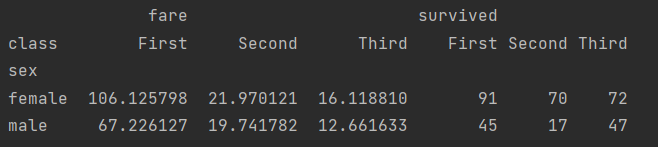
11.7 others
- Vectorization string operation
- Processing time series
- Multilevel index: used for multidimensional arrays
base_data = np.array([[1771, 11115],
[2154, 30320],
[2141, 13070],
[2424, 32680],
[1077, 7806],
[1303, 24222],
[798, 4789],
[981, 13486]])
data = pd.DataFrame(base_data, index=[
["BeiJing", "BeiJing", "ShangHai", "ShangHai", "ShenZhen", "ShenZhen", "HangZhou", "HangZhou"],
[2008, 2018] * 4], columns=["population", "GDP"])
print(data)
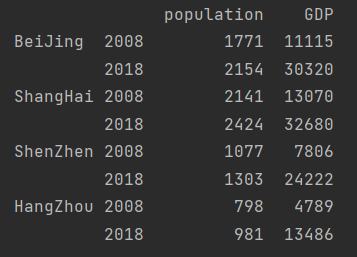
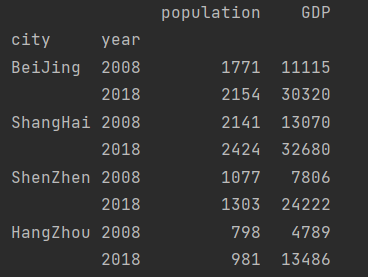
Modify row index name
data.index.names = ["city", "year"] print(data)
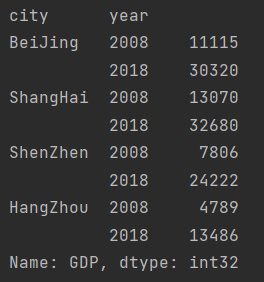
Value of column data
print(data["GDP"])
Take GDP of a city
print(data.loc["ShangHai", "GDP"])
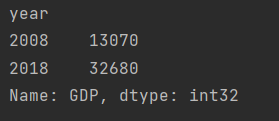
Take the situation of a city in a year
print(data.loc["ShangHai", 2018])
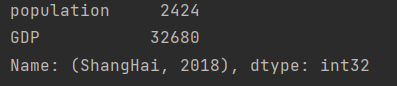
- High performance Pandas: eval()
df1, df2, df3, df4 = (pd.DataFrame(np.random.random((10000,100))) for i in range(4)) %timeit (df1+df2)/(df3+df4)
17.6 ms ± 120 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
- The memory allocation of the intermediate process of compound algebraic calculation is reduced, and the running speed is improved
%timeit pd.eval("(df1+df2)/(df3+df4)")
10.5 ms ± 153 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
It can be compared that the operation result is the same
np.allclose((df1+df2)/(df3+df4), pd.eval("(df1+df2)/(df3+df4)"))
True
- Implement inter column operation
df = pd.DataFrame(np.random.random((1000, 3)), columns=list("ABC"))
df.head()
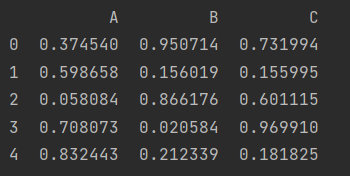
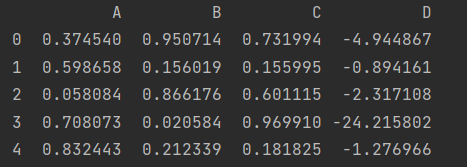
df["D"] = pd.eval("(df.A+df.B)/(df.C-1)")
df.head()
- Use local variables
column_mean = df.mean(axis=1)
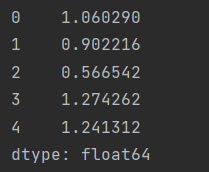
res = df.eval("A+@column_mean")
res.head()
- High performance Pandas: query()
df.head()

%timeit df[(df.A < 0.5) & (df.B > 0.5)]
1.11 ms ± 9.38 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
%timeit df.query("(A < 0.5)&(B > 0.5)")
2.55 ms ± 199 µs per loop (mean ± std. dev. of 7 runs, 100 loops each)
Instead, the time becomes longer. The reason is that the amount of data is too small, resulting in the high time cost of calling the method itself
- Use timing of eval() and query()
When the array is small, the normal method is faster
Therefore, when the amount of data is large, eval and query are called to improve the running speed
df.values.nbytes
32000
df1.values.nbytes
8000000
The above is the in-depth exploration in section 11. Pandas is an advanced tool that focuses more on data analysis.
The next section takes a closer look at the Matplotlib library.