Environmental description of this document
centos The server jupyter of scala nucleus spylon-kernel spark-2.4.0 scala-2.11.12 hadoop-2.6.0
Main contents of this paper
- spark reads the data of hive table, mainly including direct sql reading of hive table; Read hive table and hive partition table through hdfs file.
- Initialize the sparksession through the cell on the jupyter.
- At the end of the paper, there is a complete example of extracting hdfs files through spark
jupyter profile
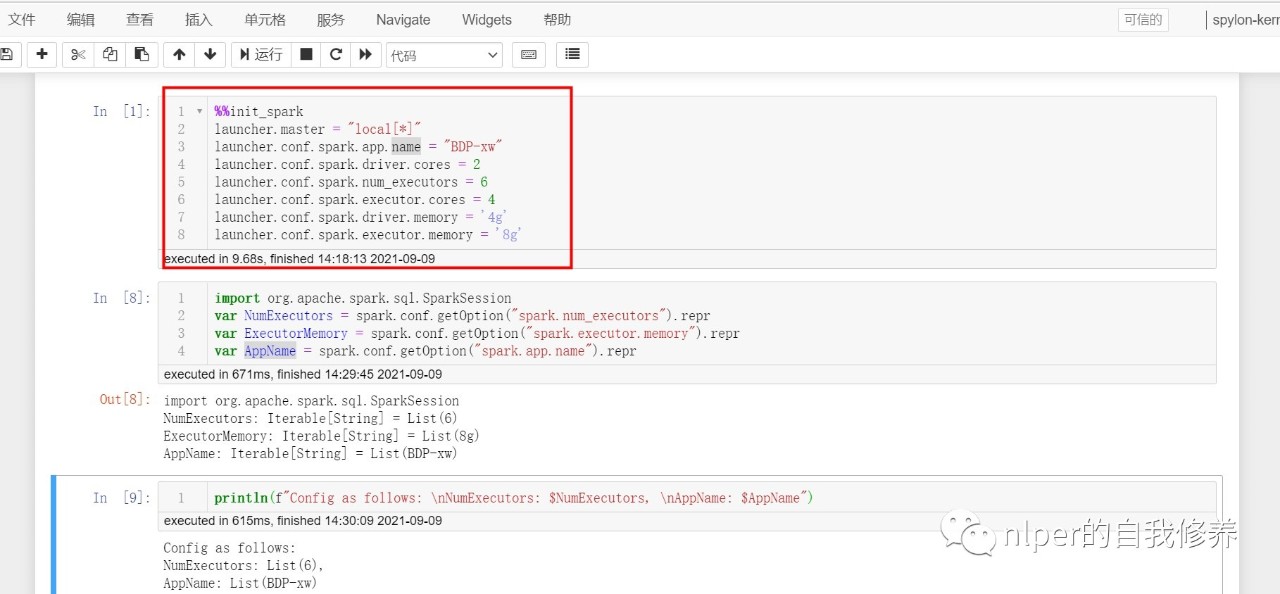
- We can initialize the spark session in the cell box of jupyter. See the following example for details.
%%init_spark launcher.master = "local[*]" launcher.conf.spark.app.name = "BDP-xw" launcher.conf.spark.driver.cores = 2 launcher.conf.spark.num_executors = 3 launcher.conf.spark.executor.cores = 4 launcher.conf.spark.driver.memory = '4g' launcher.conf.spark.executor.memory = '4g'
// launcher.conf.spark.serializer = "org.apache.spark.serializer.KryoSerializer" // launcher.conf.spark.kryoserializer.buffer.max = '4g'
import org.apache.spark.sql.SparkSession
var NumExecutors = spark.conf.getOption("spark.num_executors").repr
var ExecutorMemory = spark.conf.getOption("spark.executor.memory").repr
var AppName = spark.conf.getOption("spark.app.name").repr
var max_buffer = spark.conf.getOption("spark.kryoserializer.buffer.max").repr
println(f"Config as follows: \nNumExecutors: $NumExecutors, \nAppName: $AppName,\nmax_buffer:$max_buffer")
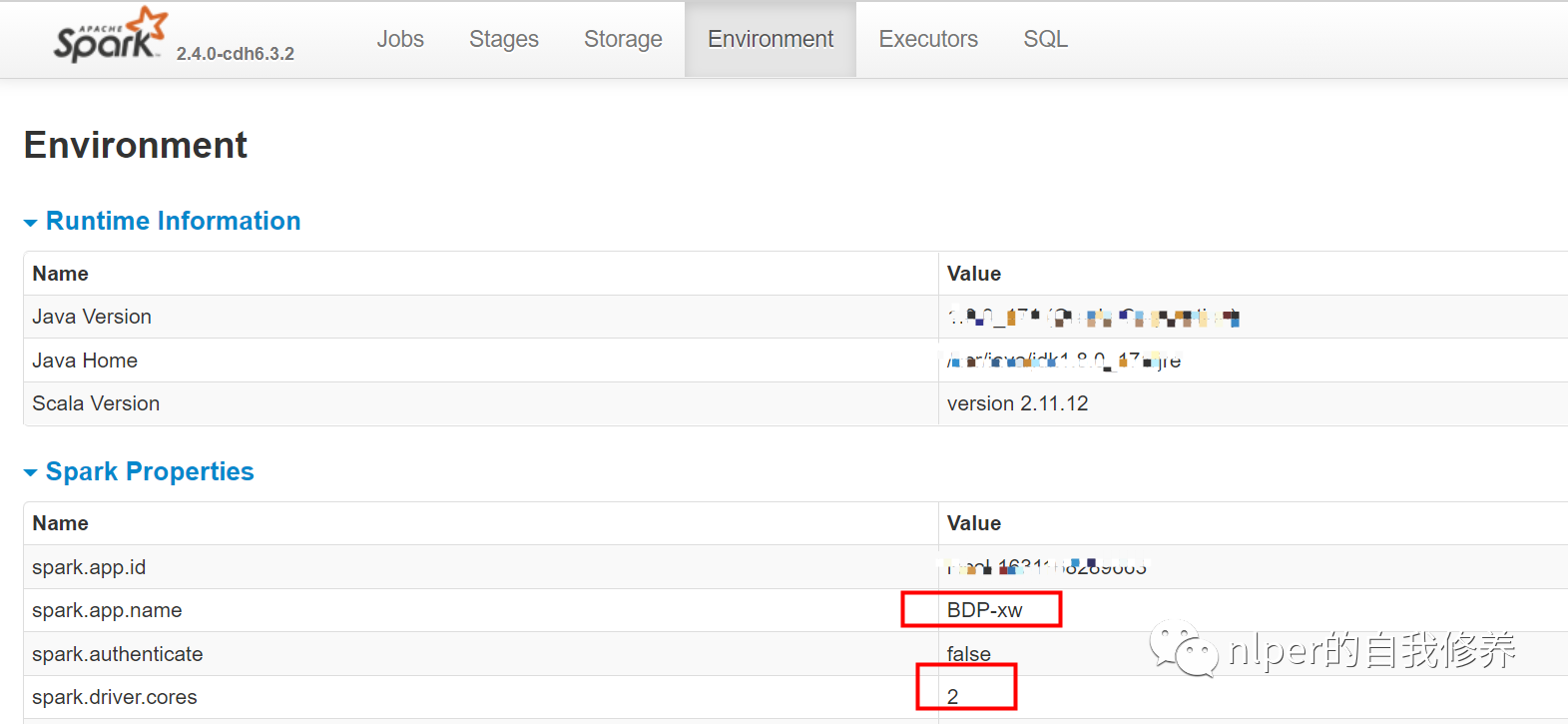
- Directly view the environment variables corresponding to our initialized sparksession
Fetch data from hive
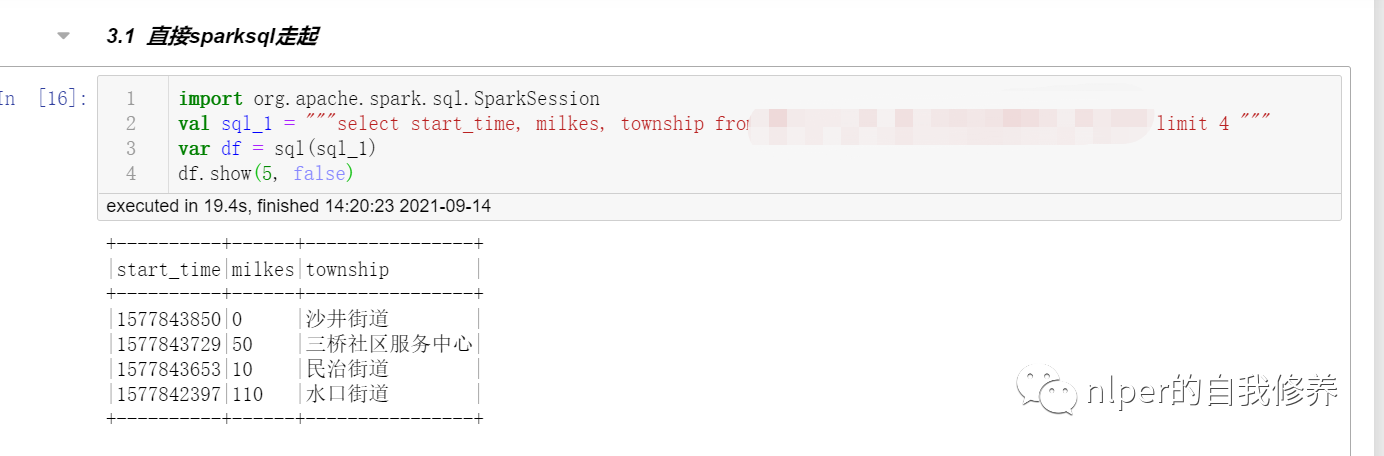
Just sparksql get up
import org.apache.spark.sql.SparkSession val sql_1 = """select * from tbs limit 4 """ var df = sql(sql_1) df.show(5, false)
Access through hdfs
- For specific examples, refer to the at the end of the article Example of a full script for fetching data from hdfs
object LoadingData_from_hdfs_base extends mylog{// with Logging
...
def main(args: Array[String]=Array("tb1", "3", "\001", "cols", "")): Unit = {
if (args.length < 2) {
println("Usage: LoadingData_from_hdfs <tb_name, parts. sep_line, cols, paths>")
System.err.println("Usage: LoadingData_from_hdfs <tb_name, parts, sep_line, cols, paths>")
System.exit(1)
}
log.warn("Here we go, dispatch")
val tb_name = args(0)
val parts = args(1)
val sep_line = args(2)
val select_col = args(3)
val save_paths = args(4)
val select_cols = select_col.split("#").toSeq
log.warn(s"Loading cols are : \n $select_cols")
val gb_sql = s"DESCRIBE FORMATTED ${tb_name}"
val gb_desc = sql(gb_sql)
val hdfs_address = gb_desc.filter($"col_name".contains("Location")).take(1)(0).getString(1)
val hdfs_address_cha = s"$hdfs_address/*/"
val Cs = new DataProcess_base(spark)
val tb_desc = Cs.get_table_desc(tb_name)
val raw_data = Cs.get_hdfs_data(hdfs_address)
val len1 = raw_data.map(item => item.split(sep_line)).first.length
val names = tb_desc.filter(!$"col_name".contains("#")).dropDuplicates(Seq("col_name")).sort("id").select("col_name").take(len1).map(_(0)).toSeq.map(_.toString)
val schema1 = StructType(names.map(fieldName => StructField(fieldName, StringType)))
val rawRDD = raw_data.map(_.split(sep_line).map(_.toString)).map(p => Row(p: _*)).filter(_.length == len1)
val df_data = spark.createDataFrame(rawRDD, schema1)//.filter("custommsgtype = '1'")
val df_desc = select_cols.toDF.join(tb_desc, $"value"===$"col_name", "left")
val df_gb_result = df_data.select(select_cols.map(df_data.col(_)): _*)//.limit(100)
df_gb_result.show(5, false)
...
// spark.stop()
}
}
val cols = "area_name#city_name#province_name" val tb_name = "tb1" val sep_line = "\u0001" // Execute script LoadingData_from_hdfs_base.main(Array(tb_name, "4", sep_line, cols, ""))
)
Determine whether the path is a folder
- Mode 1
def pathIsExist(spark: SparkSession, path: String): Boolean = {
val filePath = new org.apache.hadoop.fs.Path( path )
val fileSystem = filePath.getFileSystem( spark.sparkContext.hadoopConfiguration )
fileSystem.exists( filePath )
}
pathIsExist(spark, hdfs_address)
// The results are as follows:
// pathIsExist: (spark: org.apache.spark.sql.SparkSession, path: String)Boolean
// res4: Boolean = true
- Mode 2
import java.io.File
val d = new File("/usr/local/xw")
d.isDirectory
// The results are as follows:
// d: java.io.File = /usr/local/xw
// res3: Boolean = true
Read source data from partition table
- Attention should be paid to the partition file to ensure that there is a corresponding partition field value in the raw file on the original hdfs
- If the partition field is in the original file in hdfs, you can directly Access through hdfs
- If the partition field information is not included in the original file, you need to retrieve the data in the following way
- For specific examples, refer to the at the end of the article Example of a full script for fetching data from hdfs
Single file read
object LoadingData_from_hdfs_onefile_with_path extends mylog{
def main(args: Array[String]=Array("tb_name", "hdfs:/", "3","\n", "\001", "cols", "")): Unit = {
...
val hdfs_address = args(1)
val len1 = raw_data.map(item => item.split(sep_line)).first.length
val rawRDD = raw_data.flatMap(line => line.split(sep_text)).map(word => (word.split(sep_line):+hdfs_address)).map(p => Row(p: _*))
println(rawRDD.take(2))
val names = tb_desc.filter(!$"col_name".contains("#")).dropDuplicates(Seq("col_name")).sort("id").select("col_name").take(len1).map(_(0)).toSeq.map(_.toString)
import org.apache.spark.sql.types.StructType
val schema1 = StructType(names.map(fieldName => StructField(fieldName, StringType)))
val new_schema1 = schema1.add(StructField("path", StringType))
val df_data = spark.createDataFrame(rawRDD, new_schema1)
val df_desc = select_cols.toDF.join(tb_desc, $"value"===$"col_name", "left")
// df_desc.show(false)
val df_gb_result = df_data.select(select_cols.map(df_data.col(_)): _*)//.limit(100)
df_gb_result.show(5, false)
...
// spark.stop()
}
}
val file1 = "hdfs:file1.csv" val tb_name = "tb_name" val sep_text = "\n" val sep_line = "\001" val cols = "city#province#etl_date#path" // Execute script LoadingData_from_hdfs_onefile_with_path.main(Array(tb_name, file1, "4", sep_line, sep_text, cols, ""))
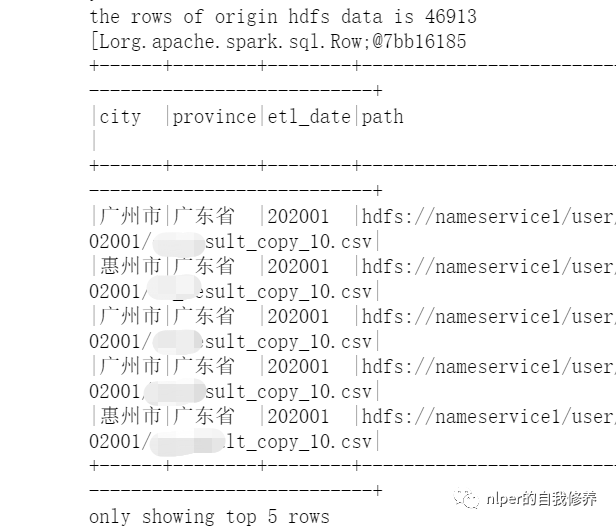
Multiple file read attempts 1
- Keep the file name information when reading the files in the folder through wholeTextFiles;
- For specific examples, refer to the at the end of the article Example of a full script for fetching data from hdfs
object LoadingData_from_hdfs_wholetext_with_path extends mylog{// with Logging
...
def main(args: Array[String]=Array("tb1", "hdfs:/", "3","\n", "\001", "cols", "")): Unit = {
...
val tb_name = args(0)
val hdfs_address = args(1)
val parts = args(2)
val sep_line = args(3)
val sep_text = args(4)
val select_col = args(5)
val save_paths = args(6)
val select_cols = select_col.split("#").toSeq
val Cs = new DataProcess_get_data(spark)
val tb_desc = Cs.get_table_desc(tb_name)
val rddWhole = spark.sparkContext.wholeTextFiles(s"$hdfs_address", 10)
rddWhole.foreach(f=>{
println(f._1+"The amount of data is=>"+f._2.split("\n").length)
})
val files = rddWhole.collect
val len1 = files.flatMap(item => item._2.split(sep_text)).take(1).flatMap(items=>items.split(sep_line)).length
val names = tb_desc.filter(!$"col_name".contains("#")).dropDuplicates(Seq("col_name")).sort("id").select("col_name").take(len1).map(_(0)).toSeq.map(_.toString)
import org.apache.spark.sql.types.StructType
// Parse the results read by wholeTextFiles and convert them into dataframe
val wordCount = files.map(f=>f._2.split(sep_text).map(g=>g.split(sep_line):+f._1.split("/").takeRight(1)(0))).flatMap(h=>h).map(p => Row(p: _*))
val schema1 = StructType(names.map(fieldName => StructField(fieldName, StringType)))
val new_schema1 = schema1.add(StructField("path", StringType))
val rawRDD = sc.parallelize(wordCount)
val df_data = spark.createDataFrame(rawRDD, new_schema1)
val df_desc = select_cols.toDF.join(tb_desc, $"value"===$"col_name", "left")
//df_desc.show(false)
val df_gb_result = df_data.select(select_cols.map(df_data.col(_)): _*)
df_gb_result.show(5, false)
println("Generated dataframe,according to path column groupby The results are as follows")
df_gb_result.groupBy("path").count().show(false)
...
// spark.stop()
}
}
val file1 = "hdfs:file1_1[01].csv" val tb_name = "tb_name" val sep_text = "\n" val sep_line = "\001" val cols = "city#province#etl_date#path" // Execute script LoadingData_from_hdfs_wholetext_with_path.main(Array(tb_name, file1, "4", sep_line, sep_text, cols, ""))
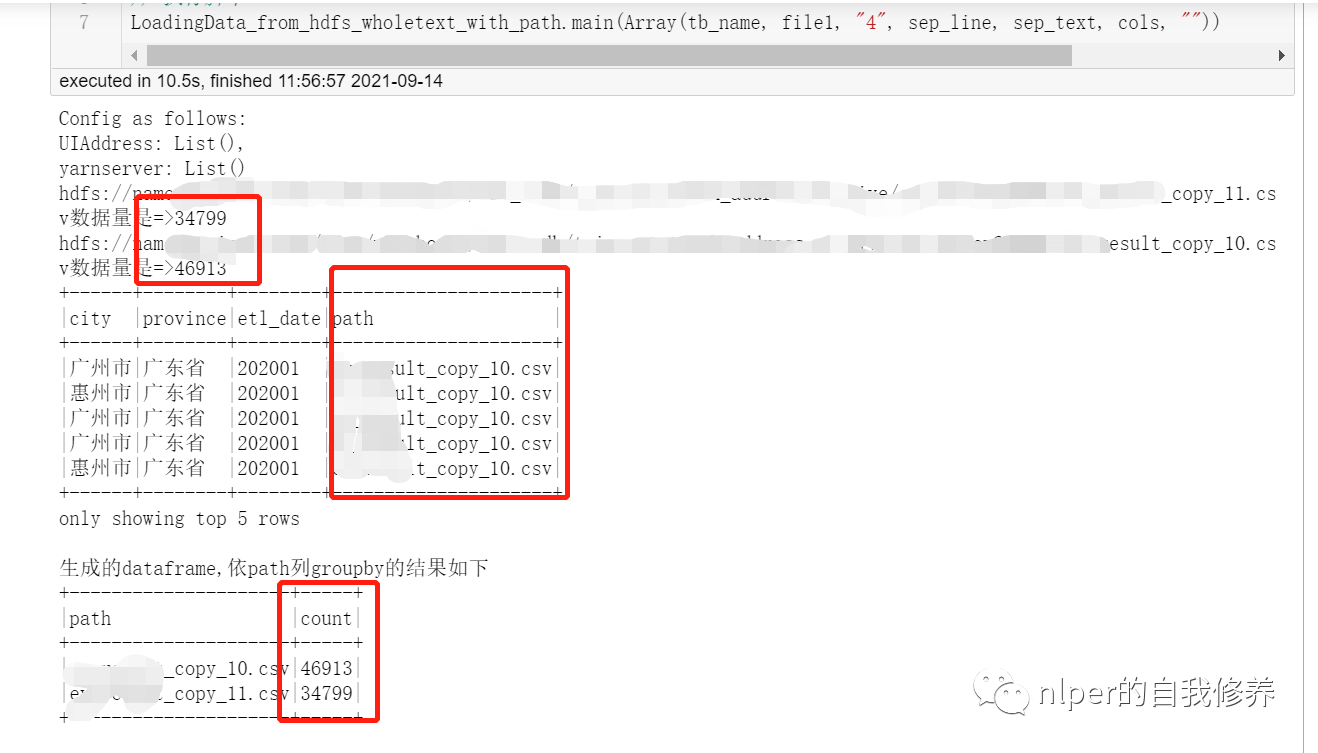
Implementation of reading multiple files and keeping file names as column names
-
The following functions are implemented:
- Split the Array[(String, String)] type by (String, String) into multiple lines;
- Divide the second element in (String, String) into multiple lines according to the \ n delimiter, and press \? The separator is divided into multiple columns;
- Add the first element in (String, String) to each line in 2. What is presented in the dataframe is a new column
-
Business scenario
- If you want to read multiple files at a time and compare them with the merged dataset, distinguish which file the data comes from.
// Test cases are mainly used to convert the results obtained from wholetextfile into DataFrame
val test1 = Array(("abasdfsdf", "a?b?c?d\nc?d?d?e"), ("sdfasdf", "b?d?a?e\nc?d?e?f"))
val test2 = test1.map(line=>line._2.split("\n").map(line1=>line1.split("\\?"):+line._1)).flatMap(line2=>line2).map(p => Row(p: _*))
val cols = "cn1#cn2#cn3#cn4#path"
val names = cols.split("#")
val schema1 = StructType(names.map(fieldName => StructField(fieldName, StringType)))
val rawRDD = sc.parallelize(test2)
val df_data = spark.createDataFrame(rawRDD, schema1)
df_data.show(4, false)
test1
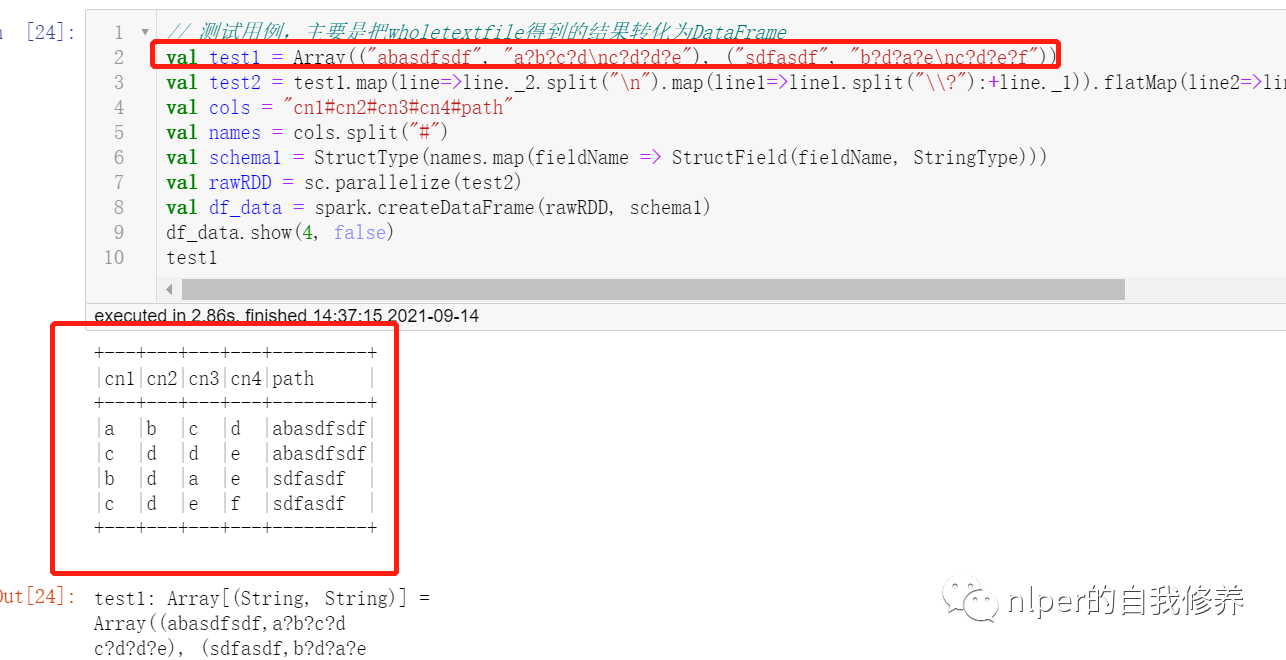
Multiple file read for loop
- Keep the file name information when reading the files in the folder through the for loop;
- For specific examples, refer to the at the end of the article Example of a full script for fetching data from hdfs
import org.apache.spark.rdd.RDD
import org.apache.spark.sql.{DataFrame, Row}
import org.apache.spark.sql.SparkSession
import org.apache.spark.{SparkConf, SparkContext}
import org.apache.spark.sql.functions.monotonically_increasing_id
import org.apache.log4j.{Level, Logger}
import org.apache.spark.sql.types.{StructType, StructField, StringType, IntegerType}
import org.apache.hadoop.fs.{FileSystem, Path}
Logger.getLogger("org").setLevel(Level.WARN)
// val log = Logger.getLogger(this.getClass)
@transient lazy val log:Logger = Logger.getLogger(this.getClass)
class DataProcess_get_data_byfor (ss: SparkSession) extends java.io.Serializable{
import ss.implicits._
import ss.sql
import org.apache.spark.sql.types.DataTypes
...
def union_dataframe(df_1:RDD[String], df_2:RDD[String]):RDD[String] ={
val count1 = df_1.map(item=>item.split(sep_line)).take(1)(0).length
val count2 = df_2.map(item=>item.split(sep_line)).take(1)(0).length
val name2 = df_2.name.split("/").takeRight(1)(0)
val arr2 = df_2.map(item=>item.split(sep_line):+name2).map(p => Row(p: _*))
println(s"That's it")
var name1 = ""
var arr1 = ss.sparkContext.makeRDD(List().map(p => Row(p: _*)))
// var arr1 = Array[org.apache.spark.sql.Row]
if (count1 == count2){
name1 = df_1.name.split("/").takeRight(1)(0)
arr1 = df_1.map(item=>item.split(sep_line):+name1).map(p => Row(p: _*))
// Arr1. Foreach (F = > Print (s "Arr1 $f" + f.length + "\n"))
println(s"Is it running here yet? $count1~$count2 $name1/$name2")
arr1
}
else{
println(s"It's running here. It's not equal, huh? $count1~$count2 $name1/$name2")
arr1 = df_1.map(item=>item.split(sep_line)).map(p => Row(p: _*))
}
var rawRDD = arr1.union(arr2)
// arr3.foreach(f=>print(s"$f" + f.length + "\n"))
// var rawRDD = sc.parallelize(arr3)
var sourceRdd = rawRDD.map(_.mkString(sep_line))
// var count31 = arr1.take(1)(0).length
// var count32 = arr2.take(1)(0).length
// var count3 = sourceRdd.map(item=>item.split(sep_line)).take(1)(0).length
// var nums = sourceRdd.count
// print(s"arr1: $count31, arr2: $count32, arr3: $count3, data amount: $nums")
sourceRdd
}
}
object LoadingData_from_hdfs_text_with_path_byfor extends mylog{// with Logging
...
def main(args: Array[String]=Array("tb1", "hdfs:/", "3","\n", "\001", "cols","data1", "test", "")): Unit = {
...
val hdfs_address = args(1)
...
val pattern = args(6)
val pattern_no = args(7)
val select_cols = select_col.split("#").toSeq
log.warn(s"Loading cols are : \n $select_cols")
val files = FileSystem.get(spark.sparkContext.hadoopConfiguration).listStatus(new Path(s"$hdfs_address"))
val files_name = files.toList.map(f=> f.getPath.getName)
val file_filter = files_name.filter(_.contains(pattern)).filterNot(_.contains(pattern_no))
val df_1 = file_filter.map(item=> sc.textFile(s"$path1$item"))
df_1.foreach(f=>{
println(f + "The amount of data is" + f.count)
})
val df2 = df_1.reduce(_ union _)
println("The amount of data after consolidation is" + df2.count)
val Cs = new DataProcess_get_data_byfor(spark)
...
// Combine the results read by the for loop
val result = df_1.reduce((a, b)=>union_dataframe(a, b))
val result2 = result.map(item=>item.split(sep_line)).map(p => Row(p: _*))
val df_data = spark.createDataFrame(result2, new_schema1)
val df_desc = select_cols.toDF.join(tb_desc, $"value"===$"col_name", "left")
println("\n")
//df_desc.show(false)
val df_gb_result = df_data.select(select_cols.map(df_data.col(_)): _*)
df_gb_result.show(5, false)
println("Generated dataframe,according to path column groupby The results are as follows")
df_gb_result.groupBy("path").count().show(false)
...
// spark.stop()
}
}
val path1 = "hdfs:202001/"
val tb_name = "tb_name"
val sep_text = "\n"
val sep_line = "\001"
val cols = "city#province#etl_date#path"
val pattern = "result_copy_1"
val pattern_no = "1.csv"
// val file_filter = List("file1_10.csv", "file_12.csv", "file_11.csv")
// Execute script LoadingData_from_hdfs_text_with_path_byfor.main(Array(tb_name, path1, "4", sep_line, sep_text, cols, pattern, pattern_no, ""))
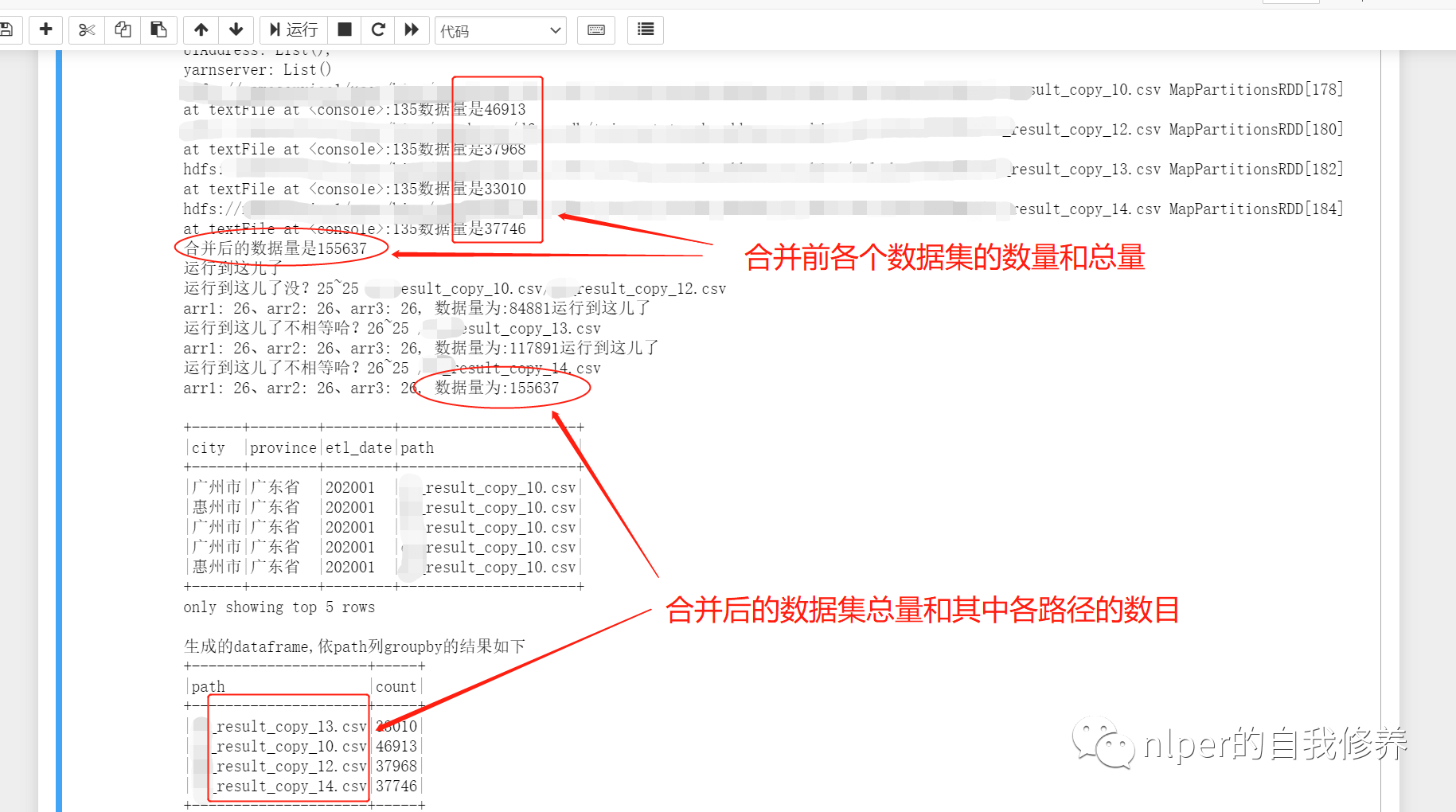
Complete example of executing script
import org.apache.spark.rdd.RDD
import org.apache.spark.sql.{DataFrame, Row}
import org.apache.spark.sql.SparkSession
import org.apache.spark.sql.functions.monotonically_increasing_id
import org.apache.log4j.{Level, Logger}
import org.apache.spark.sql.types.{StructType, StructField, StringType, IntegerType}
Logger.getLogger("org").setLevel(Level.WARN)
val log = Logger.getLogger(this.getClass)
class DataProcess_base (ss: SparkSession) extends java.io.Serializable{
import ss.implicits._
import ss.sql
import org.apache.spark.sql.types.DataTypes
def get_table_desc(tb_name:String="tb"):DataFrame ={
val gb_sql = s"desc ${tb_name}"
val gb_desc = sql(gb_sql)
val names = gb_desc.filter(!$"col_name".contains("#")).withColumn("id", monotonically_increasing_id())
names
}
def get_hdfs_data(hdfs_address:String="hdfs:"):RDD[String]={
val gb_data = ss.sparkContext.textFile(hdfs_address)
gb_data.cache()
val counts1 = gb_data.count
println(f"the rows of origin hdfs data is $counts1%-1d")
gb_data
}
}
object LoadingData_from_hdfs_base extends mylog{// with Logging
Logger.getLogger("org").setLevel(Level.WARN)
val conf = new SparkConf()
conf.setMaster("yarn")
conf.setAppName("LoadingData_From_hdfs")
conf.set("spark.home", System.getenv("SPARK_HOME"))
val spark = SparkSession.builder().config(conf).enableHiveSupport().getOrCreate()
import spark.implicits._
import spark.sql
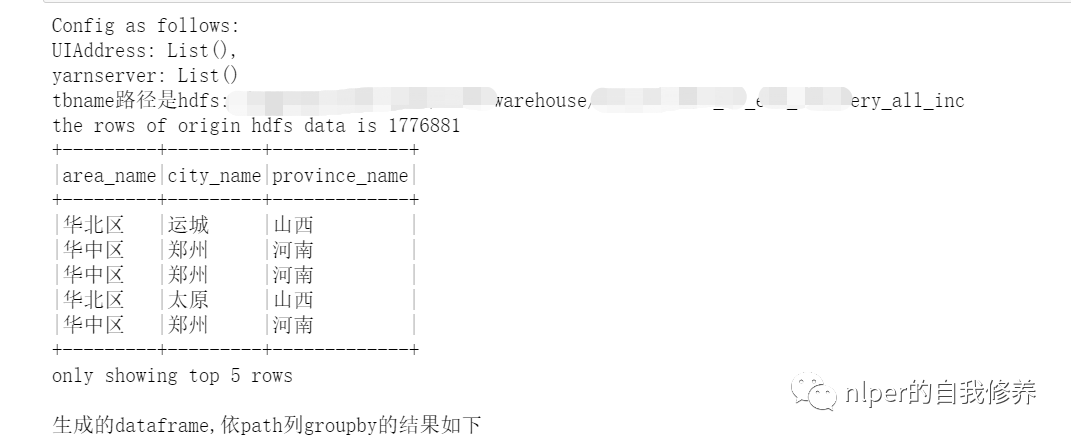
var UIAddress = spark.conf.getOption("spark.driver.appUIAddress").repr
var yarnserver = spark.conf.getOption("spark.org.apache.hadoop.yarn.server.webproxy.amfilter.AmIpFilter.param.PROXY_URI_BASES").repr
println(f"Config as follows: \nUIAddress: $UIAddress, \nyarnserver: $yarnserver")
def main(args: Array[String]=Array("tb1", "3", "\001", "cols", "")): Unit = {
if (args.length < 2) {
println("Usage: LoadingData_from_hdfs <tb_name, parts. sep_line, cols, paths>")
System.err.println("Usage: LoadingData_from_hdfs <tb_name, parts, sep_line, cols, paths>")
System.exit(1)
}
log.warn("Here we go, dispatch")
val tb_name = args(0)
val parts = args(1)
val sep_line = args(2)
val select_col = args(3)
val save_paths = args(4)
val select_cols = select_col.split("#").toSeq
log.warn(s"Loading cols are : \n $select_cols")
val gb_sql = s"DESCRIBE FORMATTED ${tb_name}"
val gb_desc = sql(gb_sql)
val hdfs_address = gb_desc.filter($"col_name".contains("Location")).take(1)(0).getString(1)
println(s"tbname Path is $hdfs_address")
val hdfs_address_cha = s"$hdfs_address/*/"
val Cs = new DataProcess_base(spark)
val tb_desc = Cs.get_table_desc(tb_name)
val raw_data = Cs.get_hdfs_data(hdfs_address)
val len1 = raw_data.map(item => item.split(sep_line)).first.length
val names = tb_desc.filter(!$"col_name".contains("#")).dropDuplicates(Seq("col_name")).sort("id").select("col_name").take(len1).map(_(0)).toSeq.map(_.toString)
val schema1 = StructType(names.map(fieldName => StructField(fieldName, StringType)))
val rawRDD = raw_data.map(_.split(sep_line).map(_.toString)).map(p => Row(p: _*)).filter(_.length == len1)
val df_data = spark.createDataFrame(rawRDD, schema1)//.filter("custommsgtype = '1'")
val df_desc = select_cols.toDF.join(tb_desc, $"value"===$"col_name", "left")
val df_gb_result = df_data.select(select_cols.map(df_data.col(_)): _*)//.limit(100)
df_gb_result.show(5, false)
println("Generated dataframe,according to path column groupby The results are as follows")
// val part = parts.toInt
// df_gb_result.repartition(part).write.mode("overwrite").option("header","true").option("sep","#").csv(save_paths)
// log.warn(f"the rows of origin data compare to mysql results is $ncounts1%-1d VS $ncounts3%-4d")
// spark.stop()
}
}
val cols = "area_name#city_name#province_name" val tb_name = "tb1" val sep_line = "\u0001" // Execute script LoadingData_from_hdfs_base.main(Array(tb_name, "4", sep_line, cols, ""))