Task01: simple graph theory and environment configuration and PyG
1, Simple graph theory
For details, please refer to datawhale open source materials
Based on the above knowledge, the brief concept of map in the field of drug discovery is summarized (to be supplemented):
Definition I (molecular diagram):
- The molecular diagram is recorded as G = { V , E } \mathcal{G}=\{\mathcal{V}, \mathcal{E}\} G={V,E}, where V = { v 1 , ... , v N } \mathcal{V}=\left\{v_{1}, \ldots, v_{N}\right\} V={v1,..., vN} is the quantity N = ∣ V ∣ N=|\mathcal{V}| N = set of atoms of ∣ V ∣, E = { e 1 , ... , e M } \mathcal{E}=\left\{e_{1}, \ldots, e_{M}\right\} E={e1,..., eM} is the quantity M M The set of chemical bonds of M.
Machine learning on molecular graph structure data
- Node prediction: the category of a prediction node or the value of a certain type of attribute
- Example: prediction of atomic type and pre training that can be used for molecular representation (such as MLM in Bert)
- Edge prediction: predicts whether there is a link between two nodes
- Examples: protein interaction, drug interaction
- Graph prediction: classify different graphs or predict the attributes of graphs
- Example: molecular attribute prediction
- Node clustering: detect whether nodes form a community
- Example: identification of functional groups (motif s)
- Other tasks
- Figure generation: for example, molecular generation
- Reverse reaction synthesis
- ......
2, Environment configuration
Install on your own computer and lab server respectively. First, check the version of pytorch installed on the computer and the version of pytorch and cudatoolkit installed on the server:
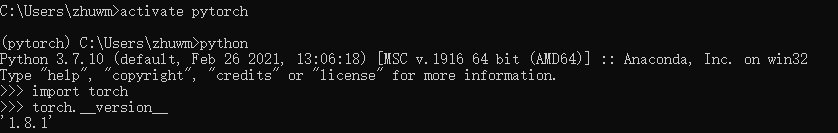
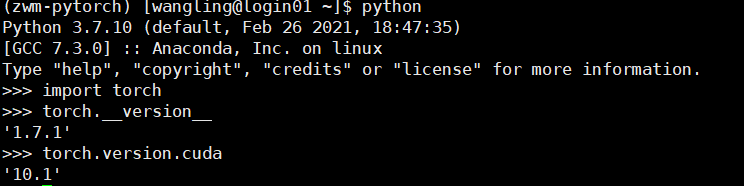
Install the correct version of PyG
# General form
pip install torch-scatter -f https://pytorch-geometric.com/whl/torch-${TORCH}+${CUDA}.html
pip install torch-sparse -f https://pytorch-geometric.com/whl/torch-${TORCH}+${CUDA}.html
pip install torch-cluster -f https://pytorch-geometric.com/whl/torch-${TORCH}+${CUDA}.html
pip install torch-spline-conv -f https://pytorch-geometric.com/whl/torch-${TORCH}+${CUDA}.html
pip install torch-geometric
# Computer installation
pip install torch-scatter -f https://pytorch-geometric.com/whl/torch-1.8.0+cpu.html
pip install torch-sparse -f https://pytorch-geometric.com/whl/torch-1.8.0+cpu.html
pip install torch-cluster -f https://pytorch-geometric.com/whl/torch-1.8.0+cpu.html
pip install torch-spline-conv -f https://pytorch-geometric.com/whl/torch-1.8.0+cpu.html
pip install torch-geometric
# Server installation
pip install torch-scatter -f https://pytorch-geometric.com/whl/torch-1.7.0+cu101.html
pip install torch-sparse -f https://pytorch-geometric.com/whl/torch-1.7.0+cu101.html
pip install torch-cluster -f https://pytorch-geometric.com/whl/torch-1.7.0+cu101.html
pip install torch-spline-conv -f https://pytorch-geometric.com/whl/torch-1.7.0+cu101.html
pip install torch-geometric
Test for successful installation:
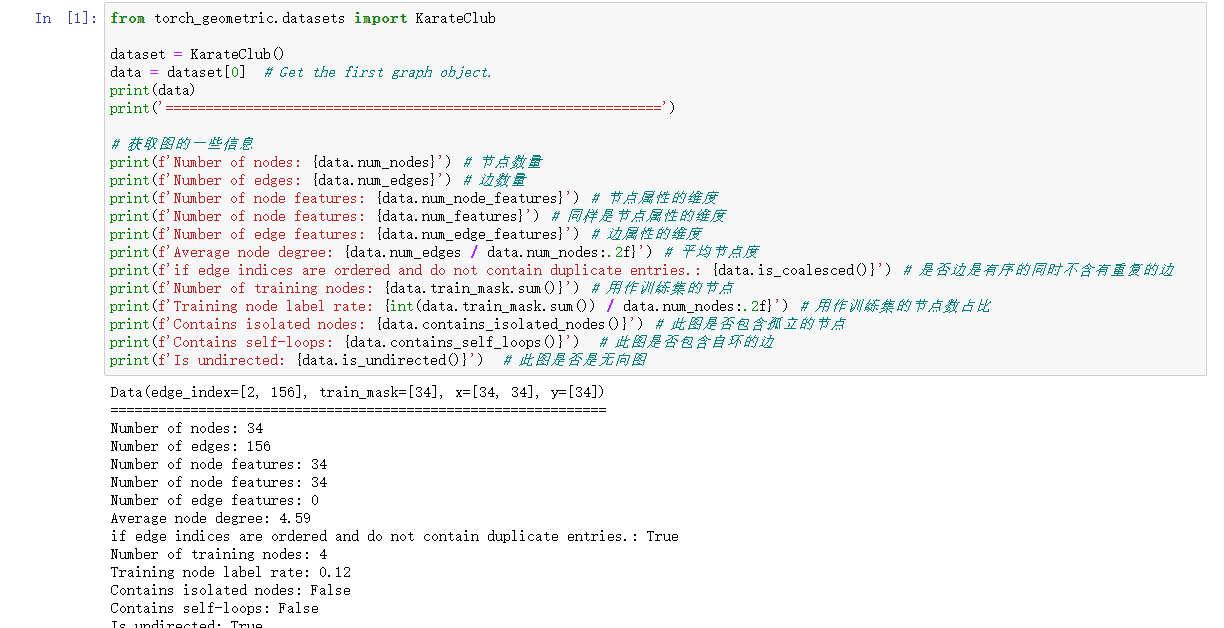
3, Representation and use of graph and graph dataset in PyG
PyTorch Geometric Library (PyG for short) is an extended library of PyTorch for geometry deep learning. Geometry deep learning refers to the deep learning applied to graphs and other irregular and unstructured data. Based on PyG library, we can easily generate a graph object according to the data, and then use it conveniently; We can also easily construct a dataset class for a graph dataset, and then easily use it for neural networks. PyG's author is Matthias Fey, and his home page is Github.
Creation of Data and Dataset objects
Here are some related links. Reading these contents will have a very intuitive understanding of Data and Dataset classes:
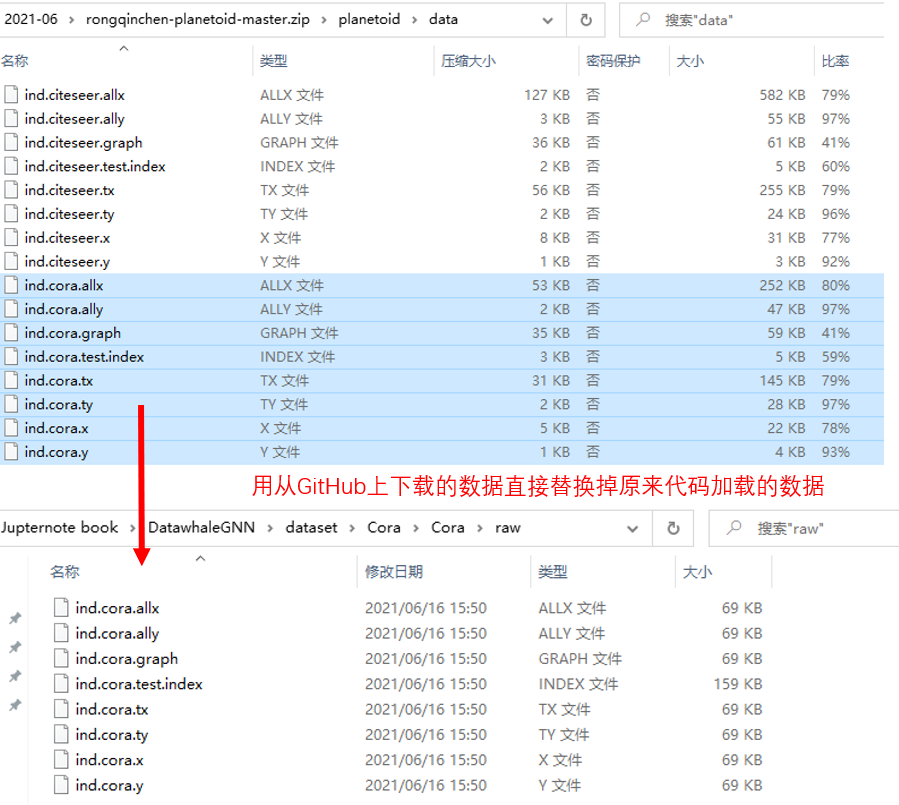
Problems encountered when downloading Cora dataset in Planetoid using datawhale open source code:
from torch_geometric.datasets import Planetoid dataset = Planetoid(root='/dataset/Cora', name='Cora') # Cora()
- Error 1: unable to access github stably due to firewall. You can solve this problem by modifying the url link of the dataset. See this article for details article.
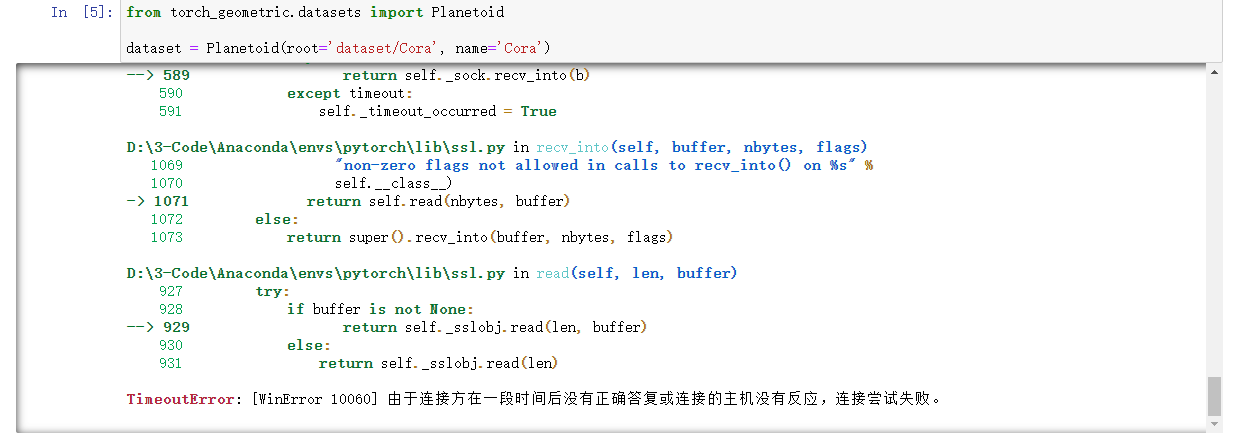
- Error 2: after reasonably changing the url, the data set can be downloaded normally, and many people in the learning group can download normally, but I made the following error:

This should be an error in the downloaded data. The final solution is to directly replace the original data with the data downloaded from github, and the data is loaded successfully.
4, Homework
- Please implement a class by inheriting the Data class, which is specially used to represent the network of "organization author paper". The network includes three types of nodes, i.e. "organization", "author" and "paper", as well as two types of edges, i.e. "author organization" and "author paper". The requirements for the classes to be implemented are: 1) storing the attributes of different nodes with different attributes; 2) storing different edges with different attributes (edges have no attributes); 3) realizing the method of obtaining the number of different nodes one by one.
The code provided by the live broadcast is as follows:
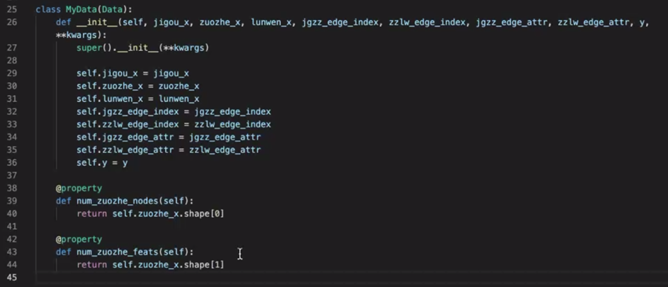
Reference others code 1,2 And sort out the following:
# Constructor of Data class:
'''
[OAP,mechanism-author-[paper]
O-Orginazation,Institutions;
A-Author,Author;
P-Paper,paper
'''
import torch
from torch_geometric.data import Data
class OAP_Data(Data):
def __init__(self, x_O=None, x_A=None, x_P=None, edge_index_A_O=None, edge_index_A_P=None, edge_attr_A_O=None, edge_attr_A_P=None, y=None, **kwargs):
r"""
Args:
x_O (Tensor, optional): Node attribute matrix, size`[num_nodes_O, num_node_O_features]`
x_A (Tensor, optional): Node attribute matrix, size`[num_nodes_A, num_node_A_features]`
x_P (Tensor, optional): Node attribute matrix, size`[num_nodes_P, num_node_P_features]`
edge_index_A_O (LongTensor, optional): Edge index matrix, size`[2, num_edges_A_O]`,The 0 line is the tail node, the 1 line is the head node, and the head points to the tail
edge_index_A_P (LongTensor, optional): Edge index matrix, size`[2, num_edges_A_P]`,The 0 line is the tail node, the 1 line is the head node, and the head points to the tail
edge_attr_A_O (Tensor, optional): Edge attribute matrix, size`[num_edges_A_O, 1]` # The edge has no attribute, so it is listed as 1
edge_attr_A_P (Tensor, optional): Edge attribute matrix, size`[num_edges_A_P, 1]` # The edge has no attribute, so it is listed as 1
y (Tensor, optional): Label of node or graph, any size (or label of edge)
"""
self.x_O = x_O # Mechanism node
self.x_A = x_A # Author class node
self.x_P = x_P # Thesis node
self.edge_index_A_O = edge_index_A_O # Author organization side serial number
self.edge_index_A_P = edge_index_A_P # Author - serial number beside the paper
# Edges have no attributes
self.edge_attr_A_O = edge_attr_A_O # Author organization edge properties
self.edge_attr_A_P = edge_attr_A_P # Author paper edge attributes
self.y = y # label
# Example method
@property
def num_nodes_O(self):
return self.x_O.shape[0] # Number of institutional nodes
@property
def num_nodes_A(self):
return self.x_A.shape[0] # Number of author nodes
@property
def num_nodes_P(self):
return self.x_P.shape[0] # Number of thesis nodes
@property
def num_edges_A_O(self):
return self.edge_index_A_O.shape[1] # Author - number of institutions
@property
def num_edges_A_P(self):
return self.edge_index_A_P.shape[1] # Author side number of papers
# Construction data: assume that the author is 3, the publishing organization is 4, and there are 5 papers in total
x_A = torch.randn(3, 6)
x_P = torch.randn(5, 7)
x_O = torch.randn(4, 5)
# Node connection relationship
edge_index_A_P = torch.tensor([
[0, 1, 2, 3, 4],
[5, 5, 5, 6, 7],
])
edge_index_A_O = torch.tensor([
[8, 9, 10, 11],
[5, 6, 7, 5],
])
# Construct a dict object
OAP_graph_dict = {
'x_O': x_O,
'x_A': x_A,
'x_P': x_P,
'edge_index_A_O': edge_index_A_O,
'edge_index_A_P': edge_index_A_P,
}
# Convert dict object to Data object
OAP_graph_data = OAP_Data.from_dict(OAP_graph_dict)
# Get the number of different nodes and edges on the OAP graph
print(f'Number of orginazation nodes: {OAP_graph_data.num_nodes_O}') # Number of nodes
print(f'Number of author nodes: {OAP_graph_data.num_nodes_A}') # Number of institutions
print(f'Number of paper nodes: {OAP_graph_data.num_nodes_P}') # Number of papers
print(f'Number of author-orginazation edges: {OAP_graph_data.num_edges_A_O}') # Author - number of institutions
print(f'Number of author-paper edges: {OAP_graph_data.num_edges_A_P}') # Author side number of papers
# output
Number of orginazation nodes: 4
Number of author nodes: 3
Number of paper nodes: 5
Number of author-orginazation edges: 4
Number of author-paper edges: 5