!git clone https://github.com/PaddleCV-SIG/PaddleSeg3D.git
1. PaddleSeg3D
From the GitHub address of this tool https://github.com/PaddleCV-SIG/PaddleSeg3D.git , it can be seen that it was developed and maintained by the CV group of the propeller interest group (PPSIG, propeller special interest group). If you are familiar with the paddle segmentation kit paddleseg, you know that paddleseg currently supports 2D segmentation and does not have 3D segmentation tools. However, many users or industries have 3D segmentation needs, especially those in the medical industry. Now the propeller interest group has developed the 3D segmentation tool of paddleseg 3D, which is expected to be integrated into the paddleseg segmentation suite later.
Now try to build a 3D segmentation project with PaddleSeg3D to segment your own medical data.
PaddleSeg3D is very similar to PaddleSeg and is easy to use, but it does not provide too many scripts for its own medical data preprocessing. For example, in the fast-running tutorial in PaddleSeg3D, the lung is segmented from the data set of new coronal chest CT. The preprocessing is based on the "measurement" of the lung, and does not support the medical format of nii suffix, So if you want to segment your data and the organ is not the lung, you need to write your own script. However, in addition to preprocessing the data, the later training is very convenient. PaddleSeg3D provides one line of code to train, verify and evaluate.
I hope PaddleSeg3D can join the PaddleSeg development kit as soon as possible.
# Installation dependency !pip install -r ./PaddleSeg3D/requirements.txt
#Decompress data !unzip -o /home/aistudio/data/data129670/livers.zip -d /home/aistudio/PaddleSeg3D
2. Data set introduction
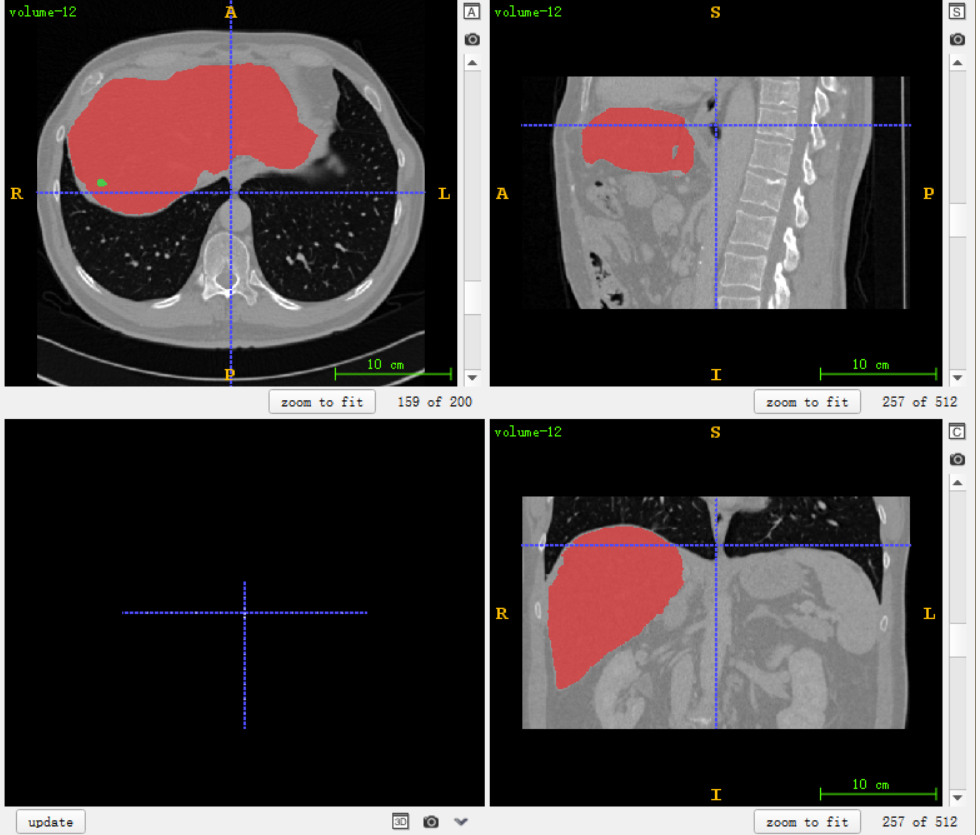
This project mainly divides the liver and tumor. The data mode is CT, which belongs to thin-layer data (voxel is about 0.7x0.7x0.7), and can reach more than 500 layers on the Z-axis. It can be seen from ITK-SNAP that the red one is the liver and the green one is the tumor. There are 28 cases in this data set
#Remember to run this line %cd /home/aistudio/PaddleSeg3D/
/home/aistudio/PaddleSeg3D
#Import common libraries import nibabel as nib import SimpleITK as sitk import matplotlib.pyplot as plt import numpy as np import os from tqdm import tqdm
3. Preprocess the data
PaddleSeg3D's quick tutorial is to segment the lungs from chest CT data. The data processing is as follows:
After reading medical data with nibabel,
if "nii.gz" in filename:
f_np = nib.load(f).get_fdata(dtype=np.float32)
Cut the CT value, resample the Size of the data to [128128], and then convert it into numpy data.
preprocess=[
HUNorm,
functools.partial(
resample, new_shape=[128, 128, 128], order=1)
],
HUNorm Default is HU_min=-1000, HU_max=600
#This value range is similar to the lung window
If you segment your own data (not the lung), you can't use the script provided by it. If your medical data is nii, it doesn't support reading directly (the script doesn't write the format of nii), and if you use nibabel to read nii, the image will rotate 90 degrees, and the clipping of CT value is also for the lung. Therefore, it is best to write your own script to preprocess your own data.
Therefore, for the liver data of this project, write your own script for the following preprocessing:
- Read with SimpleITK nii medical data will not appear with SimpleITK and will rotate 90 degrees after reading with nibabel
- Unify the direction of the data to prevent the image from turning up and down after SimpleITK reading (after unifying the direction, it will still rotate 90 degrees with nibabel)
- Cut the CT value of the data according to the window width and window level
- Resample the Size of the data to [128128]
- Convert to numpy(.npy) file
"""
find nibabel The reading will rotate 90 degrees, SImpleITK It will flip up and down. In order to solve this problem, we need to unify the direction of the data.
However, unification does not unify the direction, nibabel Will rotate 90 degrees, and PaddleSeg3D It also happens to use this database to process medical data and convert it into numpy,
So conversion numpy I don't use the script that comes with this split kit, but use it myself SImpleITK Write one.
"""
plt.figure(figsize=(10,5))
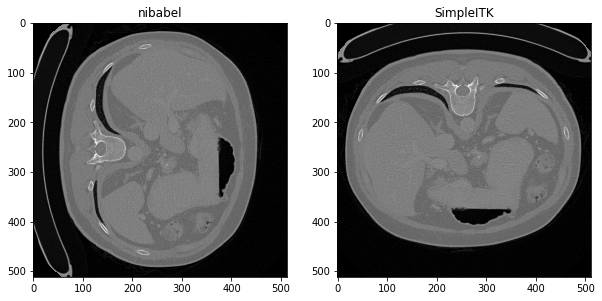
f_nib = nib.load("./livers/data/volume-2.nii").get_fdata(dtype=np.float32)
print(f"nibabel Convert file read numpy The rear shape is x,y,z{f_nib.shape}")
f_sitk= sitk.GetArrayFromImage(sitk.ReadImage('./livers/data/volume-2.nii'))
print(f"SimpleITK Read file conversion numpy The rear shape is z,y,x{f_sitk.shape}")
plt.subplot(1,2,1),plt.imshow(f_nib[:,:,60],'gray'),plt.title("nibabel")
plt.subplot(1,2,2),plt.imshow(f_sitk[60,:,:],'gray'),plt.title("SimpleITK")
plt.show()
nibabel Read file conversion numpy The rear shape is x,y,z(512, 512, 158) SimpleITK Read file conversion numpy The rear shape is z,y,x(158, 512, 512)
"""
read nii After the file, set a unified Direction,
"""
def reorient_image(image):
"""Reorients an image to standard radiology view."""
dir = np.array(image.GetDirection()).reshape(len(image.GetSize()), -1)
ind = np.argmax(np.abs(dir), axis=0)
new_size = np.array(image.GetSize())[ind]
new_spacing = np.array(image.GetSpacing())[ind]
new_extent = new_size * new_spacing
new_dir = dir[:, ind]
flip = np.diag(new_dir) < 0
flip_diag = flip * -1
flip_diag[flip_diag == 0] = 1
flip_mat = np.diag(flip_diag)
new_origin = np.array(image.GetOrigin()) + np.matmul(new_dir, (new_extent * flip))
new_dir = np.matmul(new_dir, flip_mat)
resample = sitk.ResampleImageFilter()
resample.SetOutputSpacing(new_spacing.tolist())
resample.SetSize(new_size.tolist())
resample.SetOutputDirection(new_dir.flatten().tolist())
resample.SetOutputOrigin(new_origin.tolist())
resample.SetTransform(sitk.Transform())
resample.SetDefaultPixelValue(image.GetPixelIDValue())
resample.SetInterpolator(sitk.sitkNearestNeighbor)
return resample.Execute(image)
for f in tqdm(os.listdir("./livers/data")) :
d_path = os.path.join('./livers/data',f)
seg_path = os.path.join('./livers/mask',f.replace("volume","segmentation"))
d_sitkimg = sitk.ReadImage(d_path)
d_sitkimg = reorient_image(d_sitkimg)
seg_sitkimg = sitk.ReadImage(seg_path)
seg_sitkimg = reorient_image(seg_sitkimg)
sitk.WriteImage(d_sitkimg,d_path)
sitk.WriteImage(seg_sitkimg,seg_path)
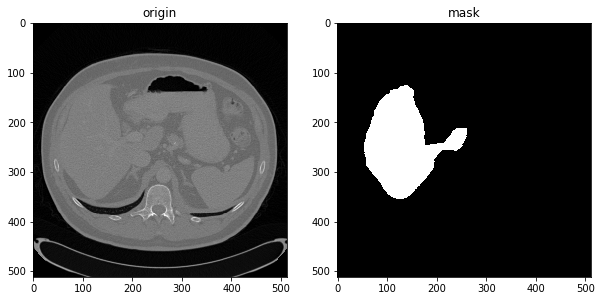
#After unifying the direction, use SimpleITK to read. It is no longer flipped up and down, and the display is normal
plt.figure(figsize=(10,5))
img= sitk.GetArrayFromImage(sitk.ReadImage('./livers/data/volume-2.nii'))
seg= sitk.GetArrayFromImage(sitk.ReadImage('./livers/mask/segmentation-2.nii'))
plt.subplot(1,2,1),plt.imshow(img[60,:,:],'gray'),plt.title("origin")
plt.subplot(1,2,2),plt.imshow(seg[60,:,:],'gray'),plt.title("mask")
plt.show()
"""
Yes, already CT Value clipping and size Convert resampled data into numpy And save.npy file
"""
from paddleseg3d.datasets.preprocess_utils import HUNorm, resample
dataset_root = "livers/"
image_dir = os.path.join(dataset_root, "images") #Do not change images. Store the npy file of the converted original data
label_dir = os.path.join(dataset_root, "labels") #Do not change labels. Store the converted label npy file
os.makedirs(image_dir, exist_ok=True)
os.makedirs(label_dir, exist_ok=True)
ww = 350
wc = 80
for f in tqdm(os.listdir("./livers/data")) :
d_path = os.path.join('./livers/data',f)
seg_path = os.path.join('./livers/mask',f.replace("volume","segmentation"))
d_img = sitk.GetArrayFromImage(sitk.ReadImage(d_path))
d_img = HUNorm(d_img,HU_min=int(wc-int(ww/2)), HU_max=int(wc+int(ww/2)))
d_img = resample(d_img,new_shape=[128, 128, 128], order=1).astype("float32")#new_shape=[z,y,x]
seg_img = sitk.GetArrayFromImage(sitk.ReadImage(seg_path))
seg_img = resample(seg_img,new_shape=[128, 128, 128], order=0).astype("int64")
np.save(os.path.join(image_dir,f.split('.')[0]), d_img)
np.save(os.path.join(label_dir,f.split('.')[0].replace("volume","segmentation")), seg_img)
# Divide the data set, and the training set and test set are 8:2
import random
random.seed(1000)
path_origin = './livers/images/'
files = list(filter(lambda x: x.endswith('.npy'), os.listdir(path_origin)))
random.shuffle(files)
rate = int(len(files) * 0.8)#Training set and test set 8:2
train_txt = open('./livers/train_list.txt','w')
val_txt = open('./livers/val_list.txt','w')
for i,f in enumerate(files):
image_path = os.path.join('images', f)
label_path = image_path.replace("images", "labels").replace('volume','segmentation')
if i < rate:
train_txt.write(image_path + ' ' + label_path+ '\n')
else:
val_txt.write(image_path + ' ' + label_path+ '\n')
train_txt.close()
val_txt.close()
print('complete')
complete
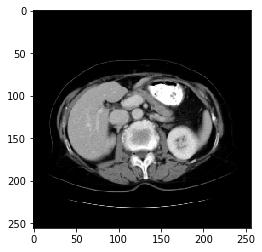
"""
Last read the converted npy File and show that the visible image shows the window width and window level of the liver, and the shape and size are also 128*128,The data are normalized to between 0 and 1
"""
data = np.load('livers/images/volume-0.npy')
print(data.shape)
print(np.max(data),np.min(data))#Normalize the data to 0-1
img = data[60,:,:]
plt.imshow(img,'gray')
plt.show()
(128, 256, 256) 1.0 0.0
4. Set training profile
PaddleSeg3D sets training parameters by configuring yaml files.
yaml files used in this training are as follows:
data_root: livers/ #You can set it as long as it is a path, because dataset_root is used to absolute path
batch_size: 2
iters: 15000
train_dataset:
type: LungCoronavirus #Never mind
dataset_root: /home/aistudio/PaddleSeg3D/livers/ #It's easy to use the absolute path here
result_dir: None #It seems useless at present
transforms:
- type: RandomResizedCrop3D #Random scale clipping
size: 128
scale: [0.8, 1.4]
- type: RandomRotation3D #Random rotation
degrees: 60 #Rotation angle [- 60, + 60] degrees
- type: RandomFlip3D # random invert
mode: train #Training mode
num_classes: 3 #Number of segmentation categories (background, liver, tumor)
val_dataset:
type: LungCoronavirus
dataset_root: /home/aistudio/PaddleSeg3D/livers/
result_dir: None
num_classes: 3
transforms: []
mode: val
optimizer: #Optimizer settings
type: sgd
momentum: 0.9
weight_decay: 1.0e-4
lr_scheduler: #Learning rate setting
type: PolynomialDecay
decay_steps: 15000
learning_rate: 0.0003
end_lr: 0
power: 0.9
loss: #Loss function, cross entropy and dice are used together
types:
- type: MixedLoss
losses:
- type: CrossEntropyLoss
weight: Null
- type: DiceLoss
coef: [1, 1] #Set the weight of cross entropy and dice
coef: [1]
model:
type: VNet #At present, there are only two 3D segmentation network options: Unet3d and Vnet
elu: False
in_channels: 1
num_classes: 3
pretrained: null
5. VNet
VNet is a medical 3D segmentation network, and its structure also adopts U-shaped structure. Compared with Unet3D, the residual structure is adopted in the layer (the first red box). Replace the pool layer with a convolution layer (second red box). The paper also emphasizes the role of Dice loss function
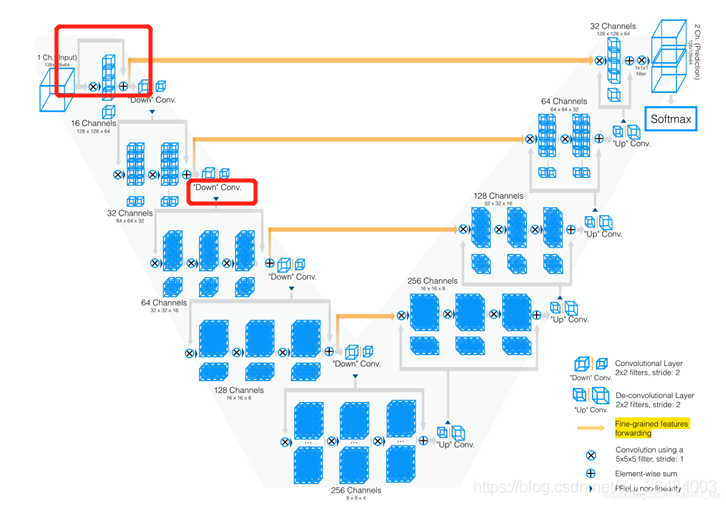
#Network structure of printing vnet !python paddleseg3d/models/vnet.py
/home/aistudio/PaddleSeg3D/paddleseg3d/models/../../paddleseg3d/cvlibs/manager.py:118: UserWarning: VNet exists already! It is now updated to <class '__main__.VNet'> !!!
component_name, component))
W0228 19:18:39.534801 27494 device_context.cc:447] Please NOTE: device: 0, GPU Compute Capability: 7.0, Driver API Version: 10.1, Runtime API Version: 10.1
W0228 19:18:39.538172 27494 device_context.cc:465] device: 0, cuDNN Version: 7.6.
/opt/conda/envs/python35-paddle120-env/lib/python3.7/site-packages/paddle/nn/layer/norm.py:653: UserWarning: When training, we now always track global mean and variance.
"When training, we now always track global mean and variance.")
out Tensor(shape=[1], dtype=float32, place=CUDAPlace(0), stop_gradient=False,
[-0.45796004]) Tensor(shape=[1], dtype=float32, place=CUDAPlace(0), stop_gradient=True,
[0.49996352])
-----------------------------------------------------------------------------------------------------------
Layer (type) Input Shape Output Shape Param #
===========================================================================================================
Conv3D-1 [[1, 1, 128, 256, 256]] [1, 16, 128, 256, 256] 2,016
BatchNorm3D-1 [[1, 16, 128, 256, 256]] [1, 16, 128, 256, 256] 64
PReLU-1 [[1, 16, 128, 256, 256]] [1, 16, 128, 256, 256] 16
InputTransition-1 [[1, 1, 128, 256, 256]] [1, 16, 128, 256, 256] 0
Conv3D-2 [[1, 16, 128, 256, 256]] [1, 32, 64, 128, 128] 4,128
BatchNorm3D-2 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 128
PReLU-2 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 32
Conv3D-3 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 128,032
BatchNorm3D-3 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 128
PReLU-4 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 32
LUConv-1 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 0
PReLU-3 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 32
DownTransition-1 [[1, 16, 128, 256, 256]] [1, 32, 64, 128, 128] 0
Conv3D-4 [[1, 32, 64, 128, 128]] [1, 64, 32, 64, 64] 16,448
BatchNorm3D-4 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 256
PReLU-5 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 64
Conv3D-5 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 512,064
BatchNorm3D-5 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 256
PReLU-7 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 64
LUConv-2 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 0
Conv3D-6 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 512,064
BatchNorm3D-6 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 256
PReLU-8 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 64
LUConv-3 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 0
PReLU-6 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 64
DownTransition-2 [[1, 32, 64, 128, 128]] [1, 64, 32, 64, 64] 0
Conv3D-7 [[1, 64, 32, 64, 64]] [1, 128, 16, 32, 32] 65,664
BatchNorm3D-7 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 512
PReLU-9 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 128
Dropout3D-3 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 0
Conv3D-8 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 2,048,128
BatchNorm3D-8 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 512
PReLU-11 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 128
LUConv-4 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 0
Conv3D-9 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 2,048,128
BatchNorm3D-9 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 512
PReLU-12 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 128
LUConv-5 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 0
Conv3D-10 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 2,048,128
BatchNorm3D-10 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 512
PReLU-13 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 128
LUConv-6 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 0
PReLU-10 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 128
DownTransition-3 [[1, 64, 32, 64, 64]] [1, 128, 16, 32, 32] 0
Conv3D-11 [[1, 128, 16, 32, 32]] [1, 256, 8, 16, 16] 262,400
BatchNorm3D-11 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 1,024
PReLU-14 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 256
Dropout3D-4 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 0
Conv3D-12 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 8,192,256
BatchNorm3D-12 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 1,024
PReLU-16 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 256
LUConv-7 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 0
Conv3D-13 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 8,192,256
BatchNorm3D-13 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 1,024
PReLU-17 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 256
LUConv-8 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 0
PReLU-15 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 256
DownTransition-4 [[1, 128, 16, 32, 32]] [1, 256, 8, 16, 16] 0
Dropout3D-5 [[1, 256, 8, 16, 16]] [1, 256, 8, 16, 16] 0
Dropout3D-6 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 0
Conv3DTranspose-1 [[1, 256, 8, 16, 16]] [1, 128, 16, 32, 32] 262,272
BatchNorm3D-14 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 512
PReLU-18 [[1, 128, 16, 32, 32]] [1, 128, 16, 32, 32] 128
Conv3D-14 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 8,192,256
BatchNorm3D-15 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 1,024
PReLU-20 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 256
LUConv-9 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 0
Conv3D-15 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 8,192,256
BatchNorm3D-16 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 1,024
PReLU-21 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 256
LUConv-10 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 0
PReLU-19 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 256
UpTransition-1 [[1, 256, 8, 16, 16], [1, 128, 16, 32, 32]] [1, 256, 16, 32, 32] 0
Dropout3D-7 [[1, 256, 16, 32, 32]] [1, 256, 16, 32, 32] 0
Dropout3D-8 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 0
Conv3DTranspose-2 [[1, 256, 16, 32, 32]] [1, 64, 32, 64, 64] 131,136
BatchNorm3D-17 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 256
PReLU-22 [[1, 64, 32, 64, 64]] [1, 64, 32, 64, 64] 64
Conv3D-16 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 2,048,128
BatchNorm3D-18 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 512
PReLU-24 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 128
LUConv-11 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 0
Conv3D-17 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 2,048,128
BatchNorm3D-19 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 512
PReLU-25 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 128
LUConv-12 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 0
PReLU-23 [[1, 128, 32, 64, 64]] [1, 128, 32, 64, 64] 128
UpTransition-2 [[1, 256, 16, 32, 32], [1, 64, 32, 64, 64]] [1, 128, 32, 64, 64] 0
Conv3DTranspose-3 [[1, 128, 32, 64, 64]] [1, 32, 64, 128, 128] 32,800
BatchNorm3D-20 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 128
PReLU-26 [[1, 32, 64, 128, 128]] [1, 32, 64, 128, 128] 32
Conv3D-18 [[1, 64, 64, 128, 128]] [1, 64, 64, 128, 128] 512,064
BatchNorm3D-21 [[1, 64, 64, 128, 128]] [1, 64, 64, 128, 128] 256
PReLU-28 [[1, 64, 64, 128, 128]] [1, 64, 64, 128, 128] 64
LUConv-13 [[1, 64, 64, 128, 128]] [1, 64, 64, 128, 128] 0
PReLU-27 [[1, 64, 64, 128, 128]] [1, 64, 64, 128, 128] 64
UpTransition-3 [[1, 128, 32, 64, 64], [1, 32, 64, 128, 128]] [1, 64, 64, 128, 128] 0
Conv3DTranspose-4 [[1, 64, 64, 128, 128]] [1, 16, 128, 256, 256] 8,208
BatchNorm3D-22 [[1, 16, 128, 256, 256]] [1, 16, 128, 256, 256] 64
PReLU-29 [[1, 16, 128, 256, 256]] [1, 16, 128, 256, 256] 16
Conv3D-19 [[1, 32, 128, 256, 256]] [1, 32, 128, 256, 256] 128,032
BatchNorm3D-23 [[1, 32, 128, 256, 256]] [1, 32, 128, 256, 256] 128
PReLU-31 [[1, 32, 128, 256, 256]] [1, 32, 128, 256, 256] 32
LUConv-14 [[1, 32, 128, 256, 256]] [1, 32, 128, 256, 256] 0
PReLU-30 [[1, 32, 128, 256, 256]] [1, 32, 128, 256, 256] 32
UpTransition-4 [[1, 64, 64, 128, 128], [1, 16, 128, 256, 256]] [1, 32, 128, 256, 256] 0
Conv3D-20 [[1, 32, 128, 256, 256]] [1, 2, 128, 256, 256] 8,002
BatchNorm3D-24 [[1, 2, 128, 256, 256]] [1, 2, 128, 256, 256] 8
PReLU-32 [[1, 2, 128, 256, 256]] [1, 2, 128, 256, 256] 2
Conv3D-21 [[1, 2, 128, 256, 256]] [1, 2, 128, 256, 256] 6
OutputTransition-1 [[1, 32, 128, 256, 256]] [1, 2, 128, 256, 256] 0
===========================================================================================================
Total params: 45,609,250
Trainable params: 45,598,618
Non-trainable params: 10,632
-----------------------------------------------------------------------------------------------------------
Input size (MB): 32.00
Forward/backward pass size (MB): 29372.00
Params size (MB): 173.99
Estimated Total Size (MB): 29577.99
-----------------------------------------------------------------------------------------------------------
Vnet test is complete
6. Start training
Start training with one line of code
#Start training
"""
!python3 train.py --config /home/aistudio/vnet_livers.yml #yaml file path
--save_dir "/home/aistudio/output/livers" #Model save path
--save_interval 500 #How many iters save model parameters
--log_iters 60 #How many iters print information once
--iters 15000 #How many iters do you train
--num_workers 6
--do_eval #Is model performance verified during training
--use_vdl #Whether to use VisualDL to visualize the trend of training indicators
"""
!python3 train.py --config /home/aistudio/vnet_livers.yml \
--save_dir "/home/aistudio/output/livers_vent128x128x128" \
--save_interval 500 --log_iters 20 \
--num_workers 6 --do_eval --use_vdl
7. Model verification and export
2022-02-28 01:24:12 [INFO] [EVAL] #Images: 6, Dice: 0.6289, Loss: 0.634533 2022-02-28 01:24:12 [INFO] [EVAL] Class dice: [0.9893 0.8679 0.0295]
After running 15000 with VNet, the effect of the validation set is as follows: the dice of liver is 0.8679 and that of tumor is 0.0295. The detection effect of these small targets is poor
#verification """ !python3 val.py --config /home/aistudio/vnet_livers.yml \ #yaml file path --model_path /home/aistudio/output/livers/best_model/model.pdparams \ #Path of optimal model --save_dir /home/aistudio/output/livers/best_model #Save path of verified results """ !python3 val.py --config /home/aistudio/vnet_livers.yml \ --model_path /home/aistudio/output/livers/best_model/model.pdparams \ --save_dir /home/aistudio/output/livers/best_model
2022-02-28 01:24:04 [INFO]
---------------Config Information---------------
batch_size: 2
data_root: livers/
iters: 15000
loss:
coef:
- 1
types:
- coef:
- 0.7
- 0.3
losses:
- type: CrossEntropyLoss
weight: null
- type: DiceLoss
type: MixedLoss
lr_scheduler:
decay_steps: 15000
end_lr: 0
learning_rate: 0.0003
power: 0.9
type: PolynomialDecay
model:
elu: false
in_channels: 1
num_classes: 3
pretrained: null
type: VNet
optimizer:
momentum: 0.9
type: sgd
weight_decay: 0.0001
train_dataset:
dataset_root: /home/aistudio/PaddleSeg3D/livers/
mode: train
num_classes: 3
result_dir: None
transforms:
- scale:
- 0.8
- 1.2
size: 128
type: RandomResizedCrop3D
- degrees: 60
type: RandomRotation3D
- type: RandomFlip3D
type: LungCoronavirus
val_dataset:
dataset_root: /home/aistudio/PaddleSeg3D/livers/
mode: val
num_classes: 3
result_dir: None
transforms: []
type: LungCoronavirus
------------------------------------------------
W0228 01:24:04.360675 32748 device_context.cc:447] Please NOTE: device: 0, GPU Compute Capability: 7.0, Driver API Version: 10.1, Runtime API Version: 10.1
W0228 01:24:04.360720 32748 device_context.cc:465] device: 0, cuDNN Version: 7.6.
2022-02-28 01:24:08 [INFO] Loading pretrained model from /home/aistudio/output/livers/best_model/model.pdparams
2022-02-28 01:24:09 [INFO] There are 178/178 variables loaded into VNet.
2022-02-28 01:24:09 [INFO] Loaded trained params of model successfully
2022-02-28 01:24:09 [INFO] Start evaluating (total_samples: 6, total_iters: 6)...
2022-02-28 01:24:10 [INFO] [EVAL] Sucessfully save iter 0 pred and label.
/opt/conda/envs/python35-paddle120-env/lib/python3.7/site-packages/paddle/fluid/dygraph/math_op_patch.py:253: UserWarning: The dtype of left and right variables are not the same, left dtype is paddle.float32, but right dtype is paddle.bool, the right dtype will convert to paddle.float32
format(lhs_dtype, rhs_dtype, lhs_dtype))
1/6 [====>.........................] - ETA: 3s - batch_cost: 0.6990 - reader cost: 0.20632022-02-28 01:24:10 [INFO] [EVAL] Sucessfully save iter 1 pred and label.
2/6 [=========>....................] - ETA: 2s - batch_cost: 0.5683 - reader cost: 0.10322022-02-28 01:24:11 [INFO] [EVAL] Sucessfully save iter 2 pred and label.
3/6 [==============>...............] - ETA: 1s - batch_cost: 0.5247 - reader cost: 0.06882022-02-28 01:24:11 [INFO] [EVAL] Sucessfully save iter 3 pred and label.
4/6 [===================>..........] - ETA: 1s - batch_cost: 0.5027 - reader cost: 0.05172022-02-28 01:24:12 [INFO] [EVAL] Sucessfully save iter 4 pred and label.
5/6 [========================>.....] - ETA: 0s - batch_cost: 0.4912 - reader cost: 0.04132022-02-28 01:24:12 [INFO] [EVAL] Sucessfully save iter 5 pred and label.
6/6 [==============================] - 3s 483ms/step - batch_cost: 0.4830 - reader cost: 0.0345
2022-02-28 01:24:12 [INFO] [EVAL] #Images: 6, Dice: 0.6289, Loss: 0.634533
2022-02-28 01:24:12 [INFO] [EVAL] Class dice:
[0.9893 0.8679 0.0295]
#Export the model to facilitate reasoning and deployment """ !python export.py --config /home/aistudio/vnet_livers.yml \ #yaml file path --save_dir '/home/aistudio/save_model' \#Save path of exported model --model_path /home/aistudio/output/livers/best_model/model.pdparams #Path of the model to be exported """ !python export.py --config /home/aistudio/vnet_livers.yml \ --save_dir '/home/aistudio/save_model' \ --model_path /home/aistudio/output/livers/best_model/model.pdparams
#Load the exported model and reason the data !python deploy/python/infer.py --config /home/aistudio/save_model/deploy.yaml \ --image_path /home/aistudio/PaddleSeg3D/livers/images/volume-5.npy \ --save_dir '/home/aistudio'
#Results after visual reasoning
pre = np.load('/home/aistudio/volume-5.npy')
pre = np.squeeze(pre)
print(pre.shape)
img = pre[60,:,:]
plt.imshow(img,'gray')
plt.show()
(256, 128, 128)
